| Full name: mesoderm posterior bHLH transcription factor 2 | Alias Symbol: SCDO2|bHLHc6 | ||
| Type: protein-coding gene | Cytoband: 15q26.1 | ||
| Entrez ID: 145873 | HGNC ID: HGNC:29659 | Ensembl Gene: ENSG00000188095 | OMIM ID: 605195 |
| Drug and gene relationship at DGIdb | |||
Expression of MESP2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MESP2 | 145873 | 1556015_a_at | 0.0028 | 0.9950 | |
| GSE26886 | MESP2 | 145873 | 1556015_a_at | -0.1952 | 0.1978 | |
| GSE45670 | MESP2 | 145873 | 1556015_a_at | 0.0685 | 0.5523 | |
| GSE53622 | MESP2 | 145873 | 36294 | -0.0770 | 0.5127 | |
| GSE53624 | MESP2 | 145873 | 36294 | -0.2770 | 0.0211 | |
| GSE63941 | MESP2 | 145873 | 1556015_a_at | 0.1582 | 0.3275 | |
| GSE77861 | MESP2 | 145873 | 1556014_at | -0.0444 | 0.7484 | |
| GSE97050 | MESP2 | 145873 | A_33_P3214463 | -0.2237 | 0.3637 | |
| TCGA | MESP2 | 145873 | RNAseq | 2.5428 | 0.0000 |
Upregulated datasets: 1; Downregulated datasets: 0.
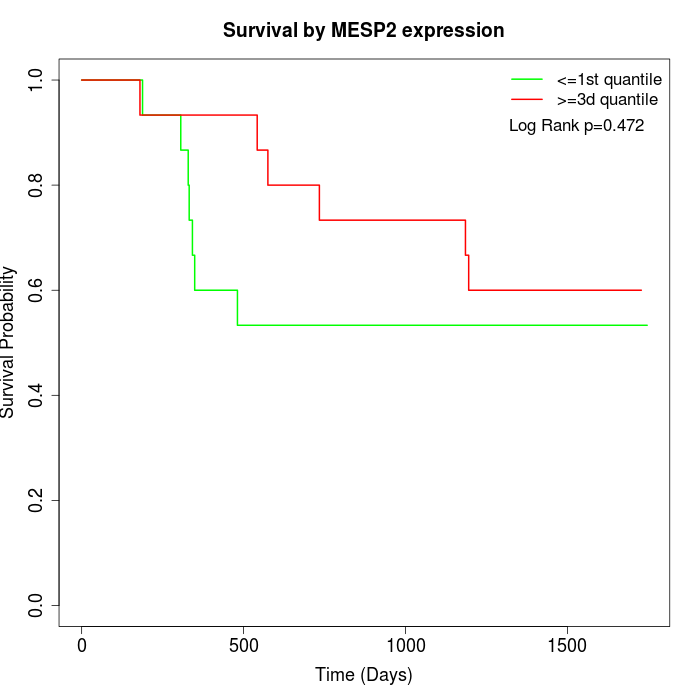
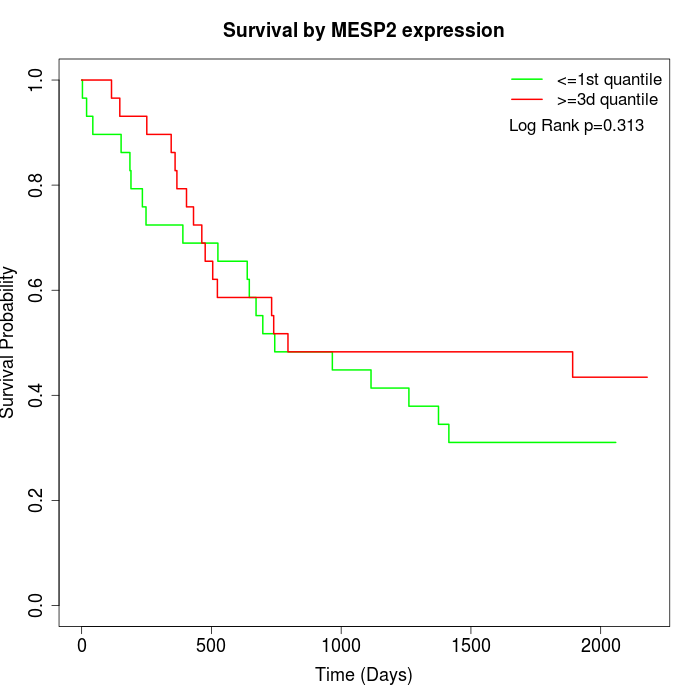
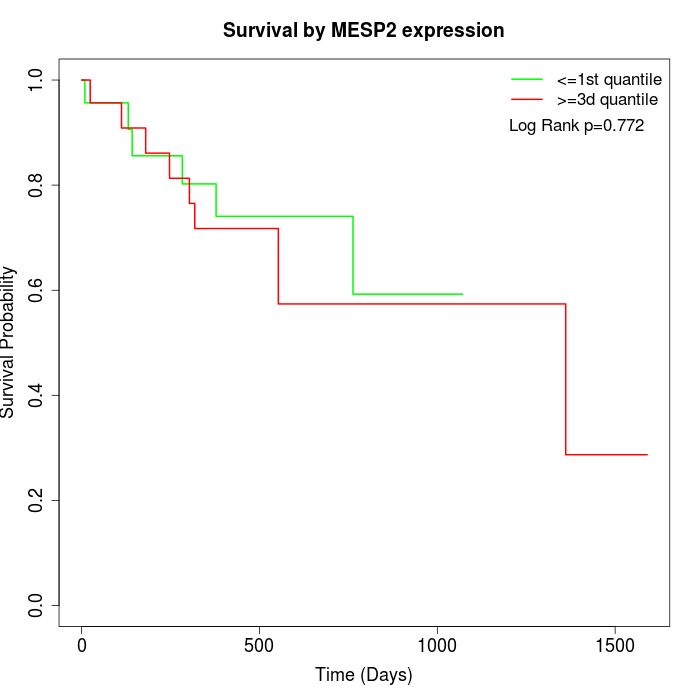
Survival by MESP2 expression:
Note: Click image to view full size file.
Copy number change of MESP2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MESP2 | 145873 | 8 | 3 | 19 | |
| GSE20123 | MESP2 | 145873 | 8 | 3 | 19 | |
| GSE43470 | MESP2 | 145873 | 5 | 4 | 34 | |
| GSE46452 | MESP2 | 145873 | 3 | 7 | 49 | |
| GSE47630 | MESP2 | 145873 | 8 | 11 | 21 | |
| GSE54993 | MESP2 | 145873 | 4 | 6 | 60 | |
| GSE54994 | MESP2 | 145873 | 7 | 6 | 40 | |
| GSE60625 | MESP2 | 145873 | 4 | 0 | 7 | |
| GSE74703 | MESP2 | 145873 | 4 | 3 | 29 | |
| GSE74704 | MESP2 | 145873 | 4 | 2 | 14 | |
| TCGA | MESP2 | 145873 | 17 | 12 | 67 |
Total number of gains: 72; Total number of losses: 57; Total Number of normals: 359.
Somatic mutations of MESP2:
Generating mutation plots.
Highly correlated genes for MESP2:
Showing top 20/55 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MESP2 | LCAT | 0.67309 | 3 | 0 | 3 |
| MESP2 | MLIP | 0.671686 | 3 | 0 | 3 |
| MESP2 | RNF208 | 0.66893 | 3 | 0 | 3 |
| MESP2 | DHX32 | 0.662158 | 3 | 0 | 3 |
| MESP2 | C6orf163 | 0.649179 | 3 | 0 | 3 |
| MESP2 | AIMP1 | 0.636792 | 3 | 0 | 3 |
| MESP2 | RBM3 | 0.635779 | 3 | 0 | 3 |
| MESP2 | NDUFA10 | 0.634856 | 4 | 0 | 3 |
| MESP2 | SAG | 0.630941 | 3 | 0 | 3 |
| MESP2 | GTF3C6 | 0.623612 | 3 | 0 | 3 |
| MESP2 | MYO3B | 0.614459 | 3 | 0 | 3 |
| MESP2 | GAL | 0.602071 | 3 | 0 | 3 |
| MESP2 | JOSD2 | 0.60148 | 3 | 0 | 3 |
| MESP2 | TMEM38A | 0.601182 | 3 | 0 | 3 |
| MESP2 | ABHD4 | 0.595709 | 5 | 0 | 3 |
| MESP2 | FAM71F2 | 0.59322 | 4 | 0 | 3 |
| MESP2 | HNRNPF | 0.591308 | 3 | 0 | 3 |
| MESP2 | KLK12 | 0.589413 | 3 | 0 | 3 |
| MESP2 | GALNT3 | 0.58656 | 4 | 0 | 3 |
| MESP2 | CERS4 | 0.584879 | 4 | 0 | 3 |
For details and further investigation, click here