| Full name: lecithin-cholesterol acyltransferase | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 16q22.1 | ||
| Entrez ID: 3931 | HGNC ID: HGNC:6522 | Ensembl Gene: ENSG00000213398 | OMIM ID: 606967 |
| Drug and gene relationship at DGIdb | |||
Expression of LCAT:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LCAT | 3931 | 204428_s_at | 0.3322 | 0.7241 | |
| GSE20347 | LCAT | 3931 | 204428_s_at | 0.2867 | 0.0104 | |
| GSE23400 | LCAT | 3931 | 204428_s_at | 0.1897 | 0.0240 | |
| GSE26886 | LCAT | 3931 | 204428_s_at | 0.2360 | 0.1941 | |
| GSE29001 | LCAT | 3931 | 204428_s_at | 0.2515 | 0.4936 | |
| GSE38129 | LCAT | 3931 | 204428_s_at | 0.0618 | 0.7837 | |
| GSE45670 | LCAT | 3931 | 204428_s_at | 0.0717 | 0.7953 | |
| GSE53622 | LCAT | 3931 | 28529 | -0.0262 | 0.8079 | |
| GSE53624 | LCAT | 3931 | 28529 | 0.0477 | 0.5844 | |
| GSE63941 | LCAT | 3931 | 204428_s_at | 0.0401 | 0.9036 | |
| GSE77861 | LCAT | 3931 | 204428_s_at | 0.1729 | 0.1376 | |
| GSE97050 | LCAT | 3931 | A_23_P218237 | -0.0654 | 0.8232 | |
| SRP064894 | LCAT | 3931 | RNAseq | 0.6768 | 0.0023 | |
| SRP133303 | LCAT | 3931 | RNAseq | -0.0711 | 0.7132 | |
| SRP193095 | LCAT | 3931 | RNAseq | 1.0046 | 0.0000 | |
| SRP219564 | LCAT | 3931 | RNAseq | -0.1857 | 0.6563 | |
| TCGA | LCAT | 3931 | RNAseq | 0.0355 | 0.7722 |
Upregulated datasets: 1; Downregulated datasets: 0.
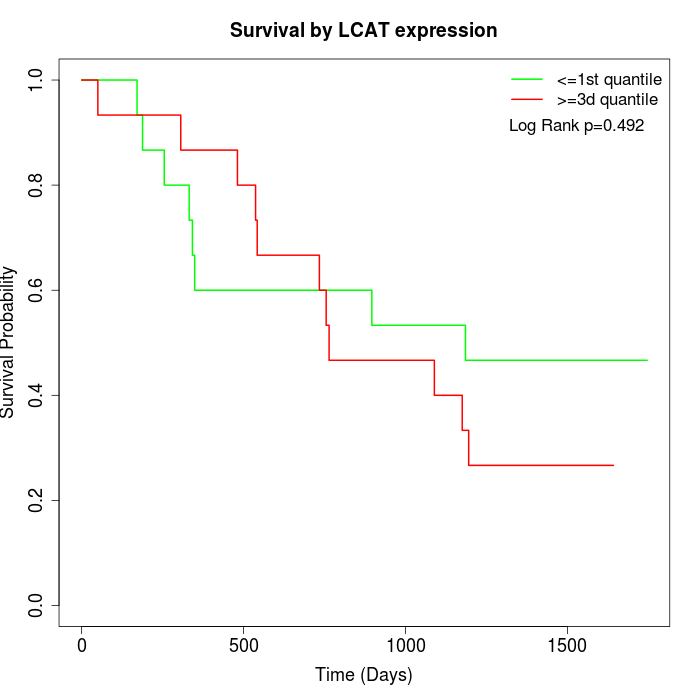
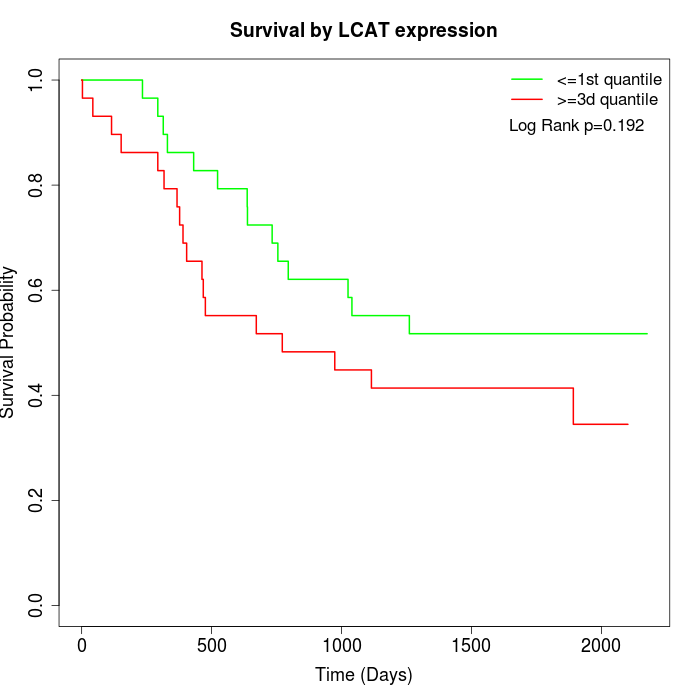
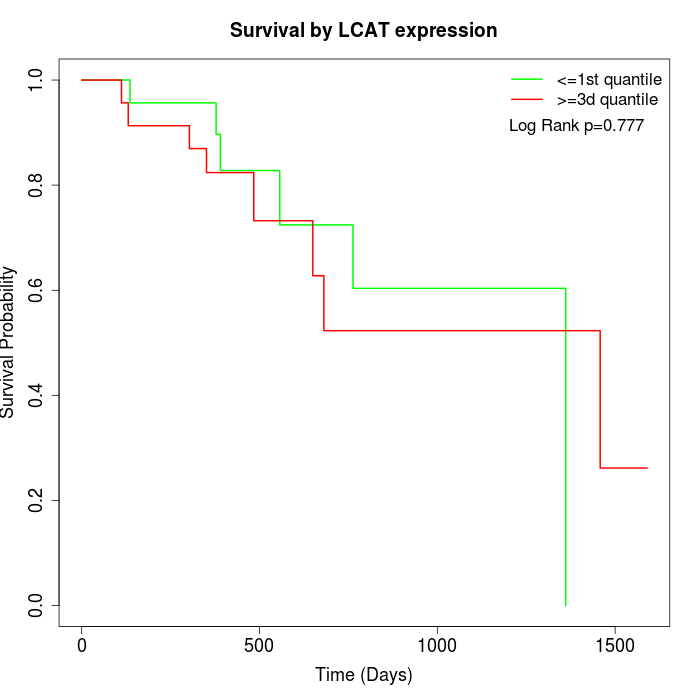
Survival by LCAT expression:
Note: Click image to view full size file.
Copy number change of LCAT:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LCAT | 3931 | 9 | 2 | 19 | |
| GSE20123 | LCAT | 3931 | 9 | 2 | 19 | |
| GSE43470 | LCAT | 3931 | 1 | 9 | 33 | |
| GSE46452 | LCAT | 3931 | 38 | 1 | 20 | |
| GSE47630 | LCAT | 3931 | 11 | 8 | 21 | |
| GSE54993 | LCAT | 3931 | 2 | 4 | 64 | |
| GSE54994 | LCAT | 3931 | 8 | 10 | 35 | |
| GSE60625 | LCAT | 3931 | 4 | 0 | 7 | |
| GSE74703 | LCAT | 3931 | 1 | 6 | 29 | |
| GSE74704 | LCAT | 3931 | 5 | 1 | 14 | |
| TCGA | LCAT | 3931 | 30 | 12 | 54 |
Total number of gains: 118; Total number of losses: 55; Total Number of normals: 315.
Somatic mutations of LCAT:
Generating mutation plots.
Highly correlated genes for LCAT:
Showing top 20/456 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LCAT | DDX59 | 0.848076 | 3 | 0 | 3 |
| LCAT | TBP | 0.843586 | 3 | 0 | 3 |
| LCAT | C9orf64 | 0.784251 | 3 | 0 | 3 |
| LCAT | KHNYN | 0.771741 | 3 | 0 | 3 |
| LCAT | ANAPC4 | 0.767558 | 3 | 0 | 3 |
| LCAT | TNFRSF10D | 0.762364 | 3 | 0 | 3 |
| LCAT | CCNB1IP1 | 0.753717 | 3 | 0 | 3 |
| LCAT | CCT5 | 0.732516 | 3 | 0 | 3 |
| LCAT | CMTM4 | 0.731141 | 3 | 0 | 3 |
| LCAT | RIMKLB | 0.72705 | 3 | 0 | 3 |
| LCAT | TRIM47 | 0.715808 | 3 | 0 | 3 |
| LCAT | TTBK2 | 0.713613 | 3 | 0 | 3 |
| LCAT | C8orf37 | 0.711527 | 3 | 0 | 3 |
| LCAT | LILRB5 | 0.710092 | 4 | 0 | 4 |
| LCAT | LIX1L | 0.704385 | 3 | 0 | 3 |
| LCAT | MAPRE3 | 0.701764 | 3 | 0 | 3 |
| LCAT | CBFA2T2 | 0.701009 | 3 | 0 | 3 |
| LCAT | GPRASP1 | 0.697775 | 3 | 0 | 3 |
| LCAT | VAMP2 | 0.693816 | 3 | 0 | 3 |
| LCAT | CHD7 | 0.693 | 3 | 0 | 3 |
For details and further investigation, click here