| Full name: mohawk homeobox | Alias Symbol: MGC39616 | ||
| Type: protein-coding gene | Cytoband: 10p12.1 | ||
| Entrez ID: 283078 | HGNC ID: HGNC:23729 | Ensembl Gene: ENSG00000150051 | OMIM ID: 601332 |
| Drug and gene relationship at DGIdb | |||
Expression of MKX:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MKX | 283078 | 239468_at | -0.3348 | 0.7175 | |
| GSE26886 | MKX | 283078 | 236014_at | 0.0961 | 0.2961 | |
| GSE45670 | MKX | 283078 | 236014_at | -0.0138 | 0.8705 | |
| GSE53622 | MKX | 283078 | 47427 | -1.7424 | 0.0000 | |
| GSE53624 | MKX | 283078 | 47427 | -1.3488 | 0.0000 | |
| GSE63941 | MKX | 283078 | 239468_at | -0.2964 | 0.8308 | |
| GSE77861 | MKX | 283078 | 236014_at | -0.0481 | 0.5551 | |
| SRP064894 | MKX | 283078 | RNAseq | -0.1396 | 0.7160 | |
| SRP133303 | MKX | 283078 | RNAseq | -1.2050 | 0.0001 | |
| SRP159526 | MKX | 283078 | RNAseq | -0.9533 | 0.0076 | |
| SRP219564 | MKX | 283078 | RNAseq | 0.0371 | 0.9732 | |
| TCGA | MKX | 283078 | RNAseq | -0.3887 | 0.2414 |
Upregulated datasets: 0; Downregulated datasets: 3.
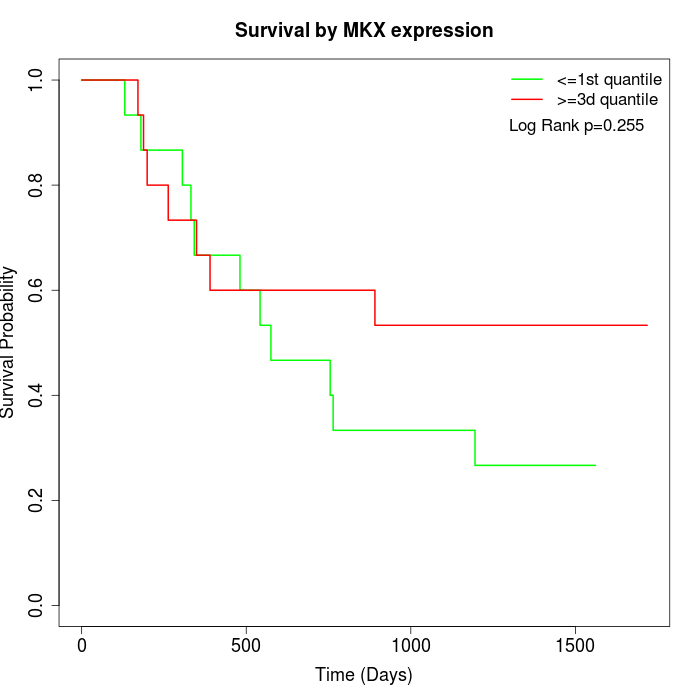
Survival by MKX expression:
Note: Click image to view full size file.
Copy number change of MKX:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MKX | 283078 | 6 | 7 | 17 | |
| GSE20123 | MKX | 283078 | 6 | 6 | 18 | |
| GSE43470 | MKX | 283078 | 3 | 4 | 36 | |
| GSE46452 | MKX | 283078 | 1 | 14 | 44 | |
| GSE47630 | MKX | 283078 | 5 | 15 | 20 | |
| GSE54993 | MKX | 283078 | 9 | 0 | 61 | |
| GSE54994 | MKX | 283078 | 3 | 9 | 41 | |
| GSE60625 | MKX | 283078 | 0 | 0 | 11 | |
| GSE74703 | MKX | 283078 | 2 | 2 | 32 | |
| GSE74704 | MKX | 283078 | 1 | 5 | 14 | |
| TCGA | MKX | 283078 | 19 | 23 | 54 |
Total number of gains: 55; Total number of losses: 85; Total Number of normals: 348.
Somatic mutations of MKX:
Generating mutation plots.
Highly correlated genes for MKX:
Showing top 20/129 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MKX | PIK3R1 | 0.739404 | 3 | 0 | 3 |
| MKX | KL | 0.730311 | 3 | 0 | 3 |
| MKX | CPED1 | 0.693253 | 3 | 0 | 3 |
| MKX | ZNF385D | 0.692905 | 3 | 0 | 3 |
| MKX | ZNF582 | 0.69057 | 3 | 0 | 3 |
| MKX | PCGF5 | 0.686491 | 3 | 0 | 3 |
| MKX | BHMT2 | 0.673887 | 3 | 0 | 3 |
| MKX | SELENBP1 | 0.669699 | 3 | 0 | 3 |
| MKX | DUSP3 | 0.669099 | 3 | 0 | 3 |
| MKX | SHISA3 | 0.656629 | 3 | 0 | 3 |
| MKX | LINC01354 | 0.655766 | 3 | 0 | 3 |
| MKX | IGSF9B | 0.655208 | 3 | 0 | 3 |
| MKX | NCAM1 | 0.654886 | 4 | 0 | 4 |
| MKX | RAI2 | 0.653691 | 3 | 0 | 3 |
| MKX | PLCL1 | 0.652077 | 4 | 0 | 3 |
| MKX | SEMA3E | 0.649511 | 3 | 0 | 3 |
| MKX | PRKAA2 | 0.649142 | 3 | 0 | 3 |
| MKX | LIMCH1 | 0.645813 | 3 | 0 | 3 |
| MKX | SGCB | 0.643306 | 3 | 0 | 3 |
| MKX | CORO2B | 0.641652 | 3 | 0 | 3 |
For details and further investigation, click here