| Full name: neurocalcin delta | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 8q22.3 | ||
| Entrez ID: 83988 | HGNC ID: HGNC:7655 | Ensembl Gene: ENSG00000104490 | OMIM ID: 606722 |
| Drug and gene relationship at DGIdb | |||
NCALD involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04740 | Olfactory transduction |
Expression of NCALD:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NCALD | 83988 | 211685_s_at | -0.6403 | 0.3817 | |
| GSE20347 | NCALD | 83988 | 211685_s_at | 0.3773 | 0.1721 | |
| GSE23400 | NCALD | 83988 | 211685_s_at | -0.1099 | 0.3975 | |
| GSE26886 | NCALD | 83988 | 211685_s_at | 0.1438 | 0.7111 | |
| GSE29001 | NCALD | 83988 | 211685_s_at | 0.2925 | 0.3125 | |
| GSE38129 | NCALD | 83988 | 211685_s_at | -0.2003 | 0.5010 | |
| GSE45670 | NCALD | 83988 | 211685_s_at | -0.7367 | 0.0107 | |
| GSE53622 | NCALD | 83988 | 47460 | -0.6084 | 0.0001 | |
| GSE53624 | NCALD | 83988 | 47460 | -0.6790 | 0.0000 | |
| GSE63941 | NCALD | 83988 | 211685_s_at | 0.2351 | 0.7435 | |
| GSE77861 | NCALD | 83988 | 211685_s_at | 0.1142 | 0.7459 | |
| GSE97050 | NCALD | 83988 | A_23_P215883 | -0.8515 | 0.3179 | |
| SRP007169 | NCALD | 83988 | RNAseq | 0.0598 | 0.9320 | |
| SRP008496 | NCALD | 83988 | RNAseq | 0.5556 | 0.2426 | |
| SRP064894 | NCALD | 83988 | RNAseq | -0.3161 | 0.3665 | |
| SRP133303 | NCALD | 83988 | RNAseq | -1.0908 | 0.0007 | |
| SRP159526 | NCALD | 83988 | RNAseq | 0.4014 | 0.3130 | |
| SRP193095 | NCALD | 83988 | RNAseq | 0.4120 | 0.0644 | |
| SRP219564 | NCALD | 83988 | RNAseq | -0.4513 | 0.6049 | |
| TCGA | NCALD | 83988 | RNAseq | -0.5576 | 0.0002 |
Upregulated datasets: 0; Downregulated datasets: 1.
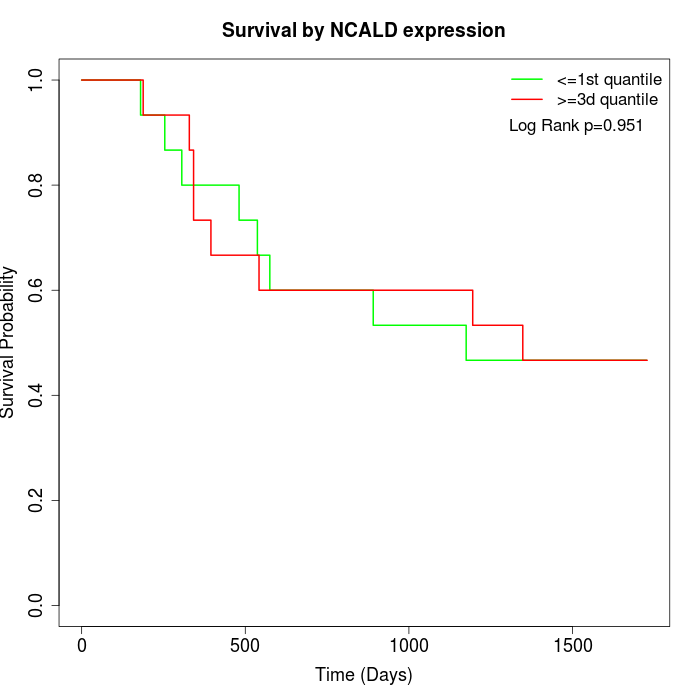
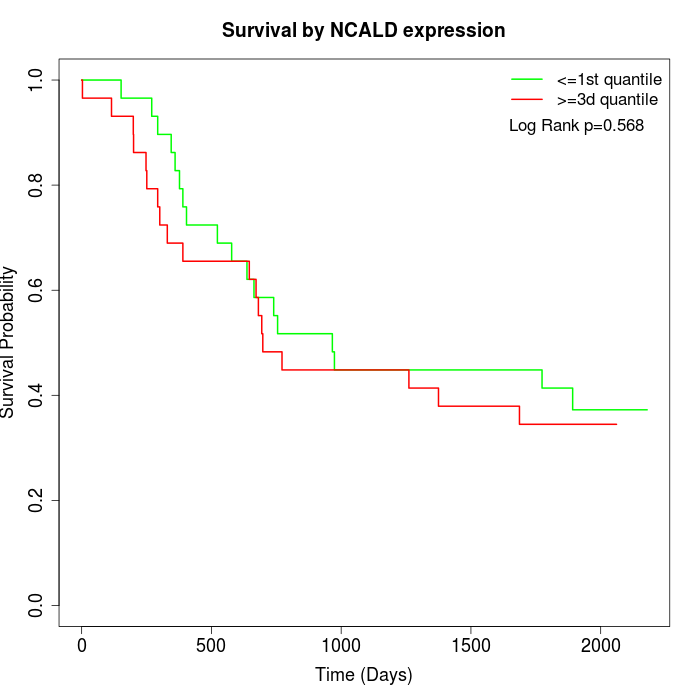
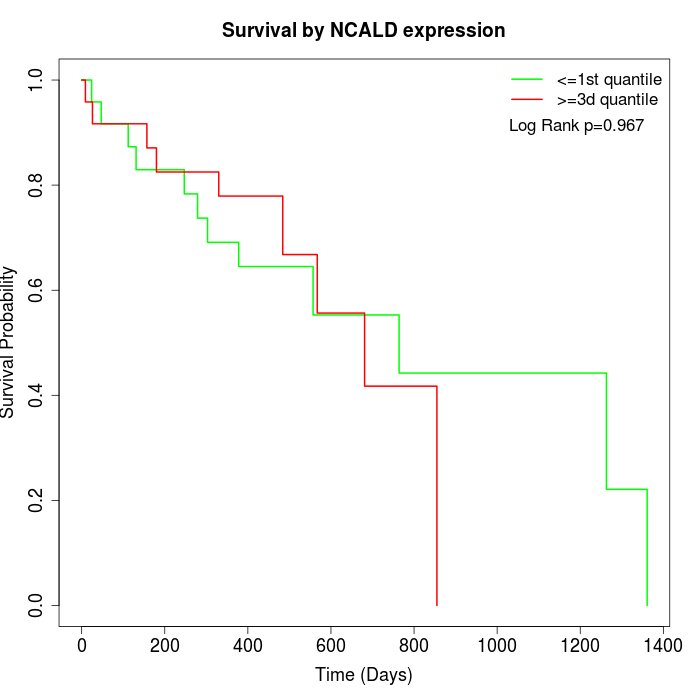
Survival by NCALD expression:
Note: Click image to view full size file.
Copy number change of NCALD:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NCALD | 83988 | 17 | 0 | 13 | |
| GSE20123 | NCALD | 83988 | 18 | 0 | 12 | |
| GSE43470 | NCALD | 83988 | 22 | 0 | 21 | |
| GSE46452 | NCALD | 83988 | 24 | 0 | 35 | |
| GSE47630 | NCALD | 83988 | 24 | 0 | 16 | |
| GSE54993 | NCALD | 83988 | 0 | 20 | 50 | |
| GSE54994 | NCALD | 83988 | 38 | 1 | 14 | |
| GSE60625 | NCALD | 83988 | 0 | 4 | 7 | |
| GSE74703 | NCALD | 83988 | 19 | 0 | 17 | |
| GSE74704 | NCALD | 83988 | 12 | 0 | 8 | |
| TCGA | NCALD | 83988 | 59 | 1 | 36 |
Total number of gains: 233; Total number of losses: 26; Total Number of normals: 229.
Somatic mutations of NCALD:
Generating mutation plots.
Highly correlated genes for NCALD:
Showing top 20/342 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NCALD | C3orf70 | 0.685714 | 4 | 0 | 4 |
| NCALD | ITGB1BP2 | 0.678207 | 5 | 0 | 5 |
| NCALD | FAM110B | 0.665045 | 4 | 0 | 3 |
| NCALD | CAPN8 | 0.656918 | 3 | 0 | 3 |
| NCALD | THAP3 | 0.653948 | 4 | 0 | 4 |
| NCALD | ARHGAP6 | 0.653759 | 6 | 0 | 6 |
| NCALD | MIR100HG | 0.651367 | 4 | 0 | 4 |
| NCALD | MYLK | 0.650963 | 7 | 0 | 7 |
| NCALD | MTFR1L | 0.649298 | 4 | 0 | 4 |
| NCALD | SLC25A4 | 0.647732 | 7 | 0 | 5 |
| NCALD | NEXN | 0.647316 | 4 | 0 | 4 |
| NCALD | NEGR1 | 0.64418 | 4 | 0 | 4 |
| NCALD | DET1 | 0.643975 | 4 | 0 | 3 |
| NCALD | CTIF | 0.643011 | 4 | 0 | 3 |
| NCALD | PENK | 0.642788 | 5 | 0 | 4 |
| NCALD | ANO5 | 0.642415 | 4 | 0 | 4 |
| NCALD | NET1 | 0.641953 | 3 | 0 | 3 |
| NCALD | CNN1 | 0.641795 | 4 | 0 | 3 |
| NCALD | MYO18B | 0.640834 | 3 | 0 | 3 |
| NCALD | CCDC8 | 0.640286 | 3 | 0 | 3 |
For details and further investigation, click here