| Full name: NIPA magnesium transporter 1 | Alias Symbol: MGC35570|SLC57A1 | ||
| Type: protein-coding gene | Cytoband: 15q11.2 | ||
| Entrez ID: 123606 | HGNC ID: HGNC:17043 | Ensembl Gene: ENSG00000170113 | OMIM ID: 608145 |
| Drug and gene relationship at DGIdb | |||
Expression of NIPA1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NIPA1 | 123606 | 225752_at | 0.6550 | 0.3815 | |
| GSE26886 | NIPA1 | 123606 | 225752_at | 0.6693 | 0.0069 | |
| GSE45670 | NIPA1 | 123606 | 225752_at | 0.3521 | 0.0279 | |
| GSE53622 | NIPA1 | 123606 | 134851 | 0.2832 | 0.0050 | |
| GSE53624 | NIPA1 | 123606 | 134851 | 0.4200 | 0.0000 | |
| GSE63941 | NIPA1 | 123606 | 225752_at | 0.2402 | 0.6426 | |
| GSE77861 | NIPA1 | 123606 | 225752_at | 1.2603 | 0.0008 | |
| GSE97050 | NIPA1 | 123606 | A_33_P3250840 | -0.1507 | 0.4921 | |
| SRP007169 | NIPA1 | 123606 | RNAseq | 2.0699 | 0.0002 | |
| SRP008496 | NIPA1 | 123606 | RNAseq | 1.4199 | 0.0002 | |
| SRP064894 | NIPA1 | 123606 | RNAseq | 0.6888 | 0.0226 | |
| SRP133303 | NIPA1 | 123606 | RNAseq | 0.7542 | 0.0005 | |
| SRP159526 | NIPA1 | 123606 | RNAseq | 0.5658 | 0.2260 | |
| SRP193095 | NIPA1 | 123606 | RNAseq | 0.3007 | 0.0759 | |
| SRP219564 | NIPA1 | 123606 | RNAseq | -0.0300 | 0.9208 | |
| TCGA | NIPA1 | 123606 | RNAseq | -0.0985 | 0.1031 |
Upregulated datasets: 3; Downregulated datasets: 0.
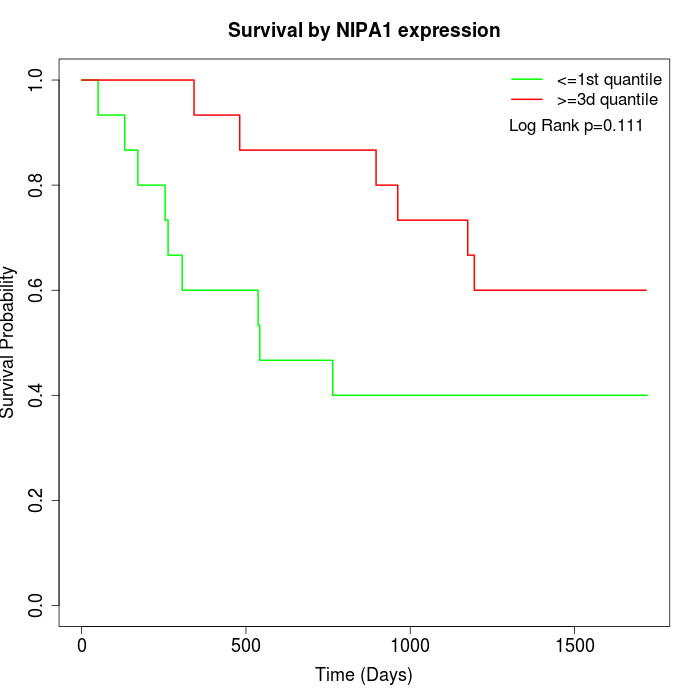
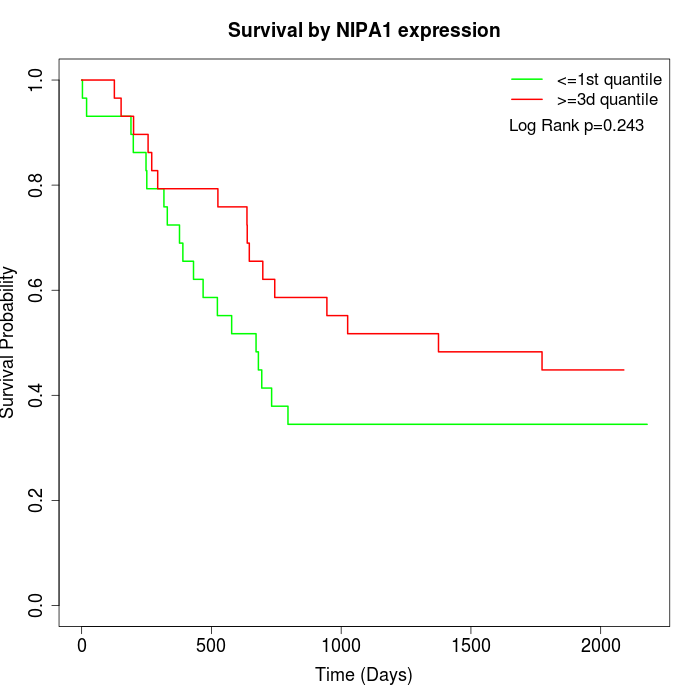
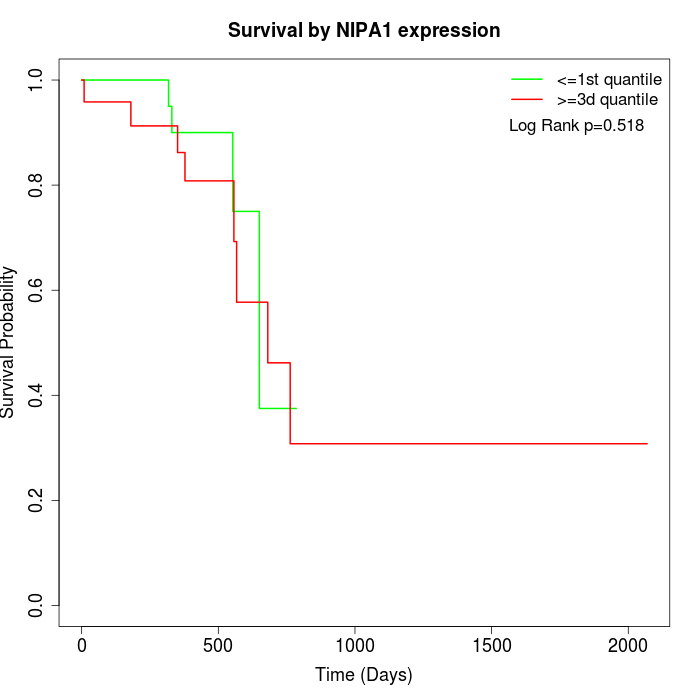
Survival by NIPA1 expression:
Note: Click image to view full size file.
Copy number change of NIPA1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NIPA1 | 123606 | 4 | 4 | 22 | |
| GSE20123 | NIPA1 | 123606 | 3 | 4 | 23 | |
| GSE43470 | NIPA1 | 123606 | 2 | 5 | 36 | |
| GSE46452 | NIPA1 | 123606 | 3 | 7 | 49 | |
| GSE47630 | NIPA1 | 123606 | 8 | 11 | 21 | |
| GSE54993 | NIPA1 | 123606 | 4 | 6 | 60 | |
| GSE54994 | NIPA1 | 123606 | 6 | 6 | 41 | |
| GSE60625 | NIPA1 | 123606 | 4 | 3 | 4 | |
| GSE74703 | NIPA1 | 123606 | 2 | 3 | 31 | |
| GSE74704 | NIPA1 | 123606 | 2 | 4 | 14 | |
| TCGA | NIPA1 | 123606 | 18 | 19 | 59 |
Total number of gains: 56; Total number of losses: 72; Total Number of normals: 360.
Somatic mutations of NIPA1:
Generating mutation plots.
Highly correlated genes for NIPA1:
Showing top 20/750 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NIPA1 | ILF3 | 0.835913 | 3 | 0 | 3 |
| NIPA1 | CPT1A | 0.797464 | 3 | 0 | 3 |
| NIPA1 | OAZ2 | 0.786293 | 3 | 0 | 3 |
| NIPA1 | UBE2Z | 0.778677 | 4 | 0 | 4 |
| NIPA1 | HSF2 | 0.777935 | 3 | 0 | 3 |
| NIPA1 | USP14 | 0.763664 | 3 | 0 | 3 |
| NIPA1 | HMG20B | 0.763581 | 3 | 0 | 3 |
| NIPA1 | EXOSC6 | 0.7632 | 3 | 0 | 3 |
| NIPA1 | PHB | 0.760836 | 4 | 0 | 4 |
| NIPA1 | TERF2 | 0.757784 | 3 | 0 | 3 |
| NIPA1 | FUS | 0.7568 | 3 | 0 | 3 |
| NIPA1 | CCT4 | 0.75171 | 4 | 0 | 4 |
| NIPA1 | COMMD2 | 0.738609 | 3 | 0 | 3 |
| NIPA1 | PDPR | 0.738581 | 3 | 0 | 3 |
| NIPA1 | GNPDA1 | 0.736998 | 3 | 0 | 3 |
| NIPA1 | AMZ2 | 0.731756 | 4 | 0 | 4 |
| NIPA1 | NUDT15 | 0.728601 | 3 | 0 | 3 |
| NIPA1 | TRMT61B | 0.728495 | 4 | 0 | 4 |
| NIPA1 | NUDT11 | 0.726602 | 3 | 0 | 3 |
| NIPA1 | TEX10 | 0.724765 | 3 | 0 | 3 |
For details and further investigation, click here