| Full name: NIPA magnesium transporter 2 | Alias Symbol: SLC57A2 | ||
| Type: protein-coding gene | Cytoband: 15q11.2 | ||
| Entrez ID: 81614 | HGNC ID: HGNC:17044 | Ensembl Gene: ENSG00000140157 | OMIM ID: 608146 |
| Drug and gene relationship at DGIdb | |||
Expression of NIPA2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NIPA2 | 81614 | 212129_at | 0.7170 | 0.1852 | |
| GSE20347 | NIPA2 | 81614 | 212129_at | 0.4695 | 0.0005 | |
| GSE23400 | NIPA2 | 81614 | 212129_at | 0.5436 | 0.0000 | |
| GSE26886 | NIPA2 | 81614 | 212129_at | 0.3722 | 0.0496 | |
| GSE29001 | NIPA2 | 81614 | 212129_at | 0.3376 | 0.1487 | |
| GSE38129 | NIPA2 | 81614 | 212129_at | 0.5479 | 0.0000 | |
| GSE45670 | NIPA2 | 81614 | 212129_at | 0.3961 | 0.0038 | |
| GSE63941 | NIPA2 | 81614 | 212129_at | 0.0158 | 0.9768 | |
| GSE77861 | NIPA2 | 81614 | 212129_at | 0.5265 | 0.0187 | |
| GSE97050 | NIPA2 | 81614 | A_33_P3250845 | 0.1477 | 0.5492 | |
| SRP007169 | NIPA2 | 81614 | RNAseq | 0.5537 | 0.2011 | |
| SRP008496 | NIPA2 | 81614 | RNAseq | 0.5914 | 0.0572 | |
| SRP064894 | NIPA2 | 81614 | RNAseq | 0.2427 | 0.1720 | |
| SRP133303 | NIPA2 | 81614 | RNAseq | 0.6201 | 0.0009 | |
| SRP159526 | NIPA2 | 81614 | RNAseq | 0.1841 | 0.6122 | |
| SRP193095 | NIPA2 | 81614 | RNAseq | 0.1433 | 0.2708 | |
| SRP219564 | NIPA2 | 81614 | RNAseq | -0.0771 | 0.7844 | |
| TCGA | NIPA2 | 81614 | RNAseq | 0.0746 | 0.1182 |
Upregulated datasets: 0; Downregulated datasets: 0.
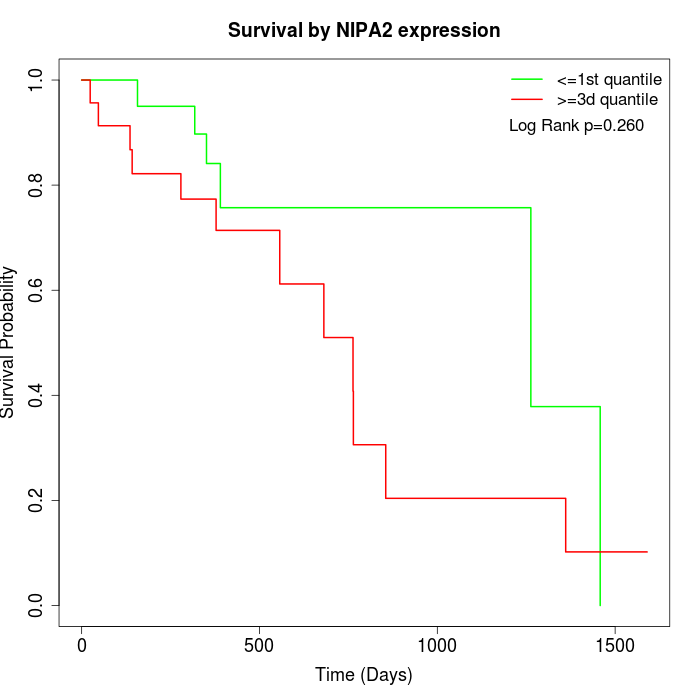
Survival by NIPA2 expression:
Note: Click image to view full size file.
Copy number change of NIPA2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NIPA2 | 81614 | 4 | 4 | 22 | |
| GSE20123 | NIPA2 | 81614 | 3 | 4 | 23 | |
| GSE43470 | NIPA2 | 81614 | 2 | 5 | 36 | |
| GSE46452 | NIPA2 | 81614 | 3 | 7 | 49 | |
| GSE47630 | NIPA2 | 81614 | 8 | 11 | 21 | |
| GSE54993 | NIPA2 | 81614 | 4 | 6 | 60 | |
| GSE54994 | NIPA2 | 81614 | 6 | 6 | 41 | |
| GSE60625 | NIPA2 | 81614 | 4 | 3 | 4 | |
| GSE74703 | NIPA2 | 81614 | 2 | 3 | 31 | |
| GSE74704 | NIPA2 | 81614 | 2 | 4 | 14 | |
| TCGA | NIPA2 | 81614 | 18 | 19 | 59 |
Total number of gains: 56; Total number of losses: 72; Total Number of normals: 360.
Somatic mutations of NIPA2:
Generating mutation plots.
Highly correlated genes for NIPA2:
Showing top 20/1448 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NIPA2 | AEBP2 | 0.817342 | 3 | 0 | 3 |
| NIPA2 | DDX55 | 0.8087 | 3 | 0 | 3 |
| NIPA2 | NUP35 | 0.799881 | 3 | 0 | 3 |
| NIPA2 | XPNPEP3 | 0.796082 | 3 | 0 | 3 |
| NIPA2 | ALG10 | 0.790382 | 3 | 0 | 3 |
| NIPA2 | SLC30A6 | 0.785091 | 4 | 0 | 4 |
| NIPA2 | RRP9 | 0.768576 | 3 | 0 | 3 |
| NIPA2 | ZBTB9 | 0.766905 | 3 | 0 | 3 |
| NIPA2 | DUS3L | 0.763276 | 3 | 0 | 3 |
| NIPA2 | CWF19L1 | 0.754418 | 4 | 0 | 4 |
| NIPA2 | TMTC3 | 0.74708 | 4 | 0 | 4 |
| NIPA2 | AJUBA | 0.746922 | 3 | 0 | 3 |
| NIPA2 | LSM4 | 0.744838 | 7 | 0 | 7 |
| NIPA2 | SAAL1 | 0.743463 | 4 | 0 | 4 |
| NIPA2 | ITPR3 | 0.742981 | 8 | 0 | 8 |
| NIPA2 | RABEP1 | 0.741325 | 3 | 0 | 3 |
| NIPA2 | WDR75 | 0.740706 | 4 | 0 | 4 |
| NIPA2 | CPSF3 | 0.740428 | 5 | 0 | 5 |
| NIPA2 | TAS2R19 | 0.739807 | 3 | 0 | 3 |
| NIPA2 | NFKB2 | 0.737523 | 3 | 0 | 3 |
For details and further investigation, click here