| Full name: glutamyl-prolyl-tRNA synthetase | Alias Symbol: EARS|PARS|GLUPRORS | ||
| Type: protein-coding gene | Cytoband: 1q41 | ||
| Entrez ID: 2058 | HGNC ID: HGNC:3418 | Ensembl Gene: ENSG00000136628 | OMIM ID: 138295 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of EPRS:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | EPRS | 2058 | 200842_s_at | 0.3049 | 0.5955 | |
| GSE20347 | EPRS | 2058 | 200842_s_at | 0.0303 | 0.8699 | |
| GSE23400 | EPRS | 2058 | 200843_s_at | 0.2074 | 0.0004 | |
| GSE26886 | EPRS | 2058 | 200843_s_at | 0.2671 | 0.2317 | |
| GSE29001 | EPRS | 2058 | 200842_s_at | 0.0610 | 0.8695 | |
| GSE38129 | EPRS | 2058 | 200842_s_at | 0.2340 | 0.0765 | |
| GSE45670 | EPRS | 2058 | 200843_s_at | 0.2586 | 0.1110 | |
| GSE53622 | EPRS | 2058 | 37028 | 0.2670 | 0.0000 | |
| GSE53624 | EPRS | 2058 | 37028 | 0.2646 | 0.0000 | |
| GSE63941 | EPRS | 2058 | 200843_s_at | -0.0534 | 0.8920 | |
| GSE77861 | EPRS | 2058 | 200843_s_at | 0.4144 | 0.0425 | |
| GSE97050 | EPRS | 2058 | A_23_P97632 | 0.2007 | 0.4164 |
Upregulated datasets: 0; Downregulated datasets: 0.
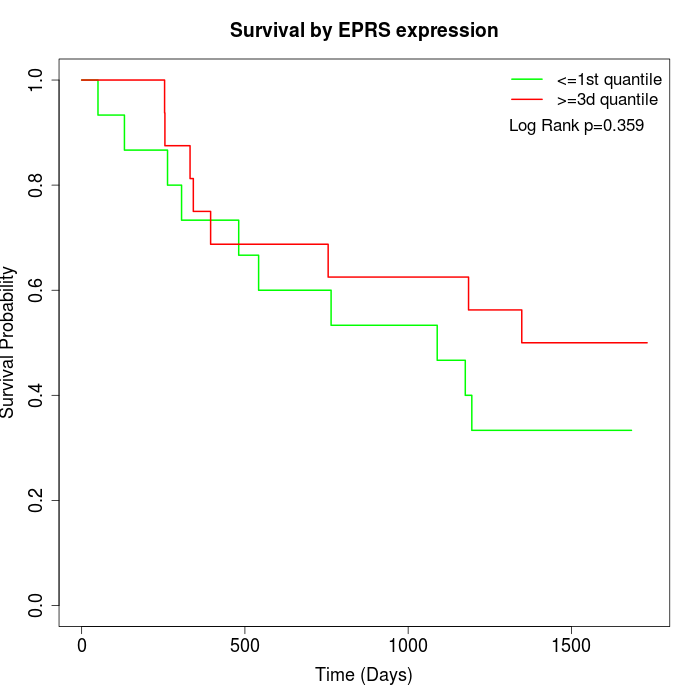
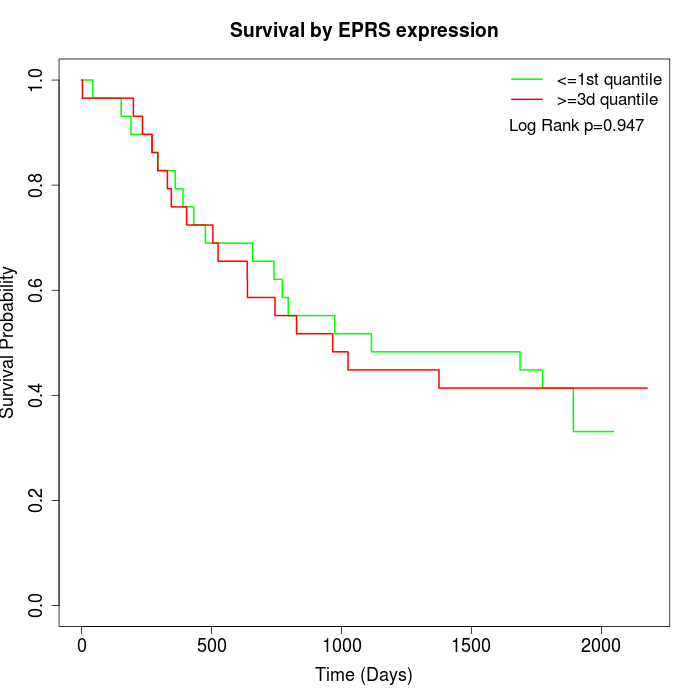
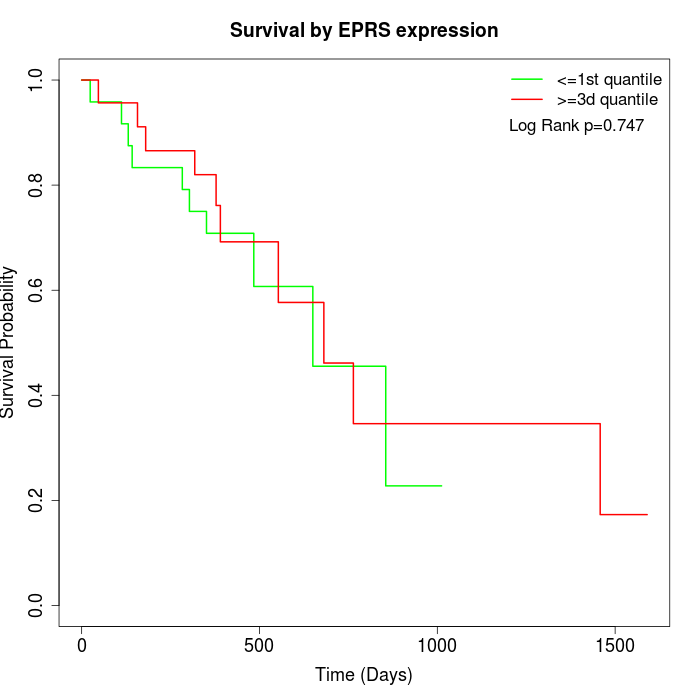
Survival by EPRS expression:
Note: Click image to view full size file.
Copy number change of EPRS:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | EPRS | 2058 | 9 | 0 | 21 | |
| GSE20123 | EPRS | 2058 | 8 | 0 | 22 | |
| GSE43470 | EPRS | 2058 | 8 | 1 | 34 | |
| GSE46452 | EPRS | 2058 | 3 | 1 | 55 | |
| GSE47630 | EPRS | 2058 | 15 | 0 | 25 | |
| GSE54993 | EPRS | 2058 | 0 | 6 | 64 | |
| GSE54994 | EPRS | 2058 | 15 | 0 | 38 | |
| GSE60625 | EPRS | 2058 | 0 | 0 | 11 | |
| GSE74703 | EPRS | 2058 | 8 | 1 | 27 | |
| GSE74704 | EPRS | 2058 | 2 | 0 | 18 | |
| TCGA | EPRS | 2058 | 46 | 3 | 47 |
Total number of gains: 114; Total number of losses: 12; Total Number of normals: 362.
Somatic mutations of EPRS:
Generating mutation plots.
Highly correlated genes for EPRS:
Showing top 20/765 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| EPRS | NIPBL | 0.756074 | 3 | 0 | 3 |
| EPRS | ABCD4 | 0.753129 | 3 | 0 | 3 |
| EPRS | TYW5 | 0.748863 | 3 | 0 | 3 |
| EPRS | DDX20 | 0.748424 | 3 | 0 | 3 |
| EPRS | SNRNP200 | 0.740715 | 3 | 0 | 3 |
| EPRS | STX16 | 0.72095 | 3 | 0 | 3 |
| EPRS | SS18L1 | 0.717334 | 4 | 0 | 4 |
| EPRS | ASH1L | 0.714758 | 3 | 0 | 3 |
| EPRS | RNF13 | 0.711994 | 3 | 0 | 3 |
| EPRS | ALKBH3 | 0.707858 | 4 | 0 | 3 |
| EPRS | ANAPC1 | 0.707687 | 3 | 0 | 3 |
| EPRS | SPRED1 | 0.705359 | 3 | 0 | 3 |
| EPRS | MAP3K7 | 0.705137 | 4 | 0 | 3 |
| EPRS | PDPR | 0.702237 | 3 | 0 | 3 |
| EPRS | SESN3 | 0.701616 | 3 | 0 | 3 |
| EPRS | TADA1 | 0.700987 | 3 | 0 | 3 |
| EPRS | CTBP2 | 0.698376 | 3 | 0 | 3 |
| EPRS | TOMM40L | 0.697427 | 3 | 0 | 3 |
| EPRS | CHD7 | 0.693549 | 4 | 0 | 3 |
| EPRS | VGLL4 | 0.689957 | 4 | 0 | 3 |
For details and further investigation, click here