| Full name: NK2 homeobox 3 | Alias Symbol: NKX2.3|CSX3|NKX4-3 | ||
| Type: protein-coding gene | Cytoband: 10q24.2 | ||
| Entrez ID: 159296 | HGNC ID: HGNC:7836 | Ensembl Gene: ENSG00000119919 | OMIM ID: 606727 |
| Drug and gene relationship at DGIdb | |||
Expression of NKX2-3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NKX2-3 | 159296 | 1553808_a_at | -0.0354 | 0.9200 | |
| GSE26886 | NKX2-3 | 159296 | 1553808_a_at | 0.2241 | 0.3720 | |
| GSE45670 | NKX2-3 | 159296 | 1553808_a_at | -0.0706 | 0.8538 | |
| GSE53622 | NKX2-3 | 159296 | 44215 | -0.3025 | 0.0001 | |
| GSE53624 | NKX2-3 | 159296 | 44215 | -0.4444 | 0.0000 | |
| GSE63941 | NKX2-3 | 159296 | 1553808_a_at | 0.3706 | 0.3937 | |
| GSE77861 | NKX2-3 | 159296 | 1553808_a_at | 0.0207 | 0.8614 | |
| GSE97050 | NKX2-3 | 159296 | A_24_P38702 | 0.1284 | 0.6513 | |
| SRP159526 | NKX2-3 | 159296 | RNAseq | -0.2408 | 0.8253 | |
| TCGA | NKX2-3 | 159296 | RNAseq | -1.4653 | 0.0029 |
Upregulated datasets: 0; Downregulated datasets: 1.
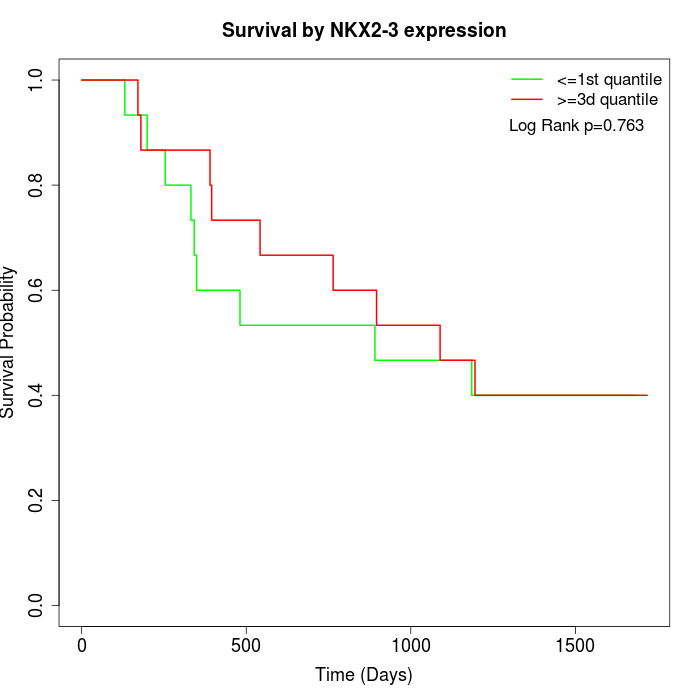
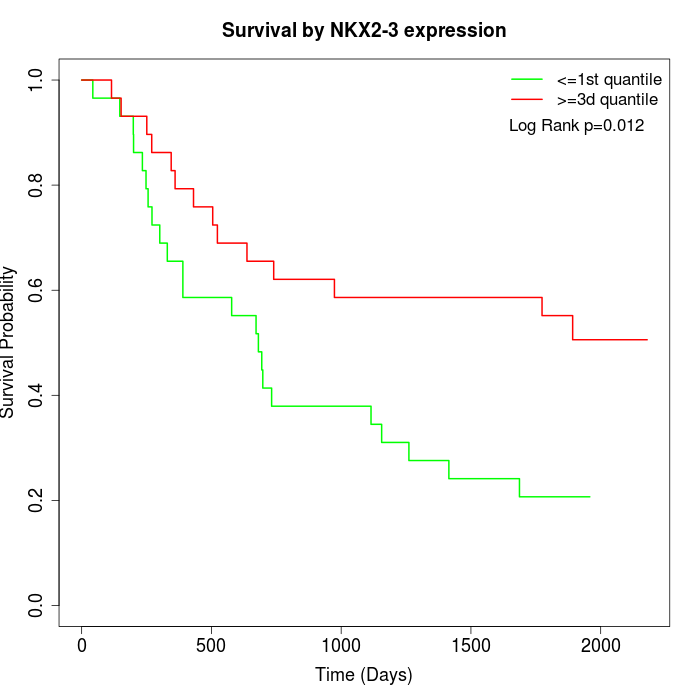
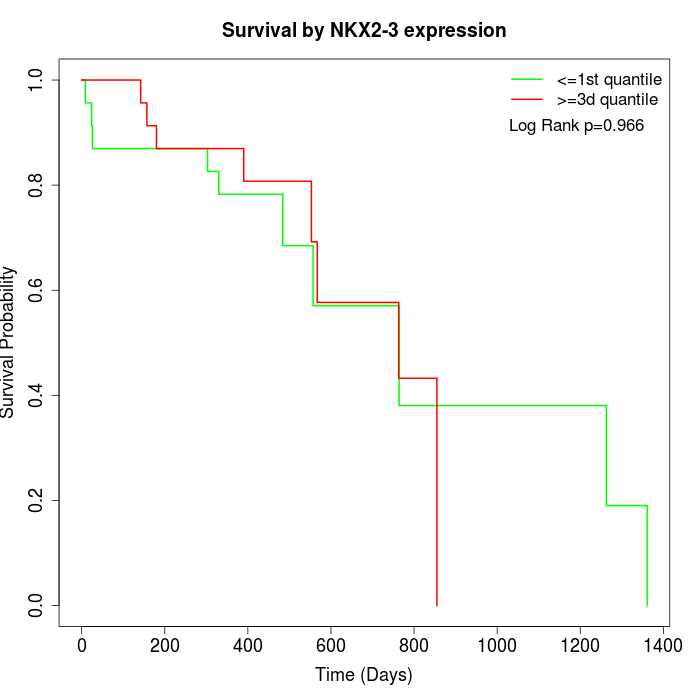
Survival by NKX2-3 expression:
Note: Click image to view full size file.
Copy number change of NKX2-3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NKX2-3 | 159296 | 1 | 8 | 21 | |
| GSE20123 | NKX2-3 | 159296 | 1 | 7 | 22 | |
| GSE43470 | NKX2-3 | 159296 | 0 | 7 | 36 | |
| GSE46452 | NKX2-3 | 159296 | 0 | 11 | 48 | |
| GSE47630 | NKX2-3 | 159296 | 2 | 14 | 24 | |
| GSE54993 | NKX2-3 | 159296 | 8 | 0 | 62 | |
| GSE54994 | NKX2-3 | 159296 | 1 | 12 | 40 | |
| GSE60625 | NKX2-3 | 159296 | 0 | 0 | 11 | |
| GSE74703 | NKX2-3 | 159296 | 0 | 5 | 31 | |
| GSE74704 | NKX2-3 | 159296 | 0 | 4 | 16 |
Total number of gains: 13; Total number of losses: 68; Total Number of normals: 311.
Somatic mutations of NKX2-3:
Generating mutation plots.
Highly correlated genes for NKX2-3:
Showing top 20/51 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NKX2-3 | KIRREL2 | 0.754637 | 3 | 0 | 3 |
| NKX2-3 | SLC22A31 | 0.749101 | 3 | 0 | 3 |
| NKX2-3 | KCNH3 | 0.74648 | 3 | 0 | 3 |
| NKX2-3 | HTR6 | 0.723551 | 3 | 0 | 3 |
| NKX2-3 | GDF7 | 0.718538 | 3 | 0 | 3 |
| NKX2-3 | SLC22A12 | 0.714641 | 3 | 0 | 3 |
| NKX2-3 | FOXA3 | 0.711773 | 4 | 0 | 4 |
| NKX2-3 | RUFY4 | 0.705459 | 3 | 0 | 3 |
| NKX2-3 | TMEM125 | 0.704206 | 3 | 0 | 3 |
| NKX2-3 | RHBDL1 | 0.702145 | 3 | 0 | 3 |
| NKX2-3 | KBTBD13 | 0.7012 | 3 | 0 | 3 |
| NKX2-3 | GH1 | 0.687611 | 4 | 0 | 3 |
| NKX2-3 | PKD2L1 | 0.682218 | 3 | 0 | 3 |
| NKX2-3 | ZC3H10 | 0.676283 | 5 | 0 | 4 |
| NKX2-3 | CLDN24 | 0.67581 | 3 | 0 | 3 |
| NKX2-3 | MFSD2B | 0.675797 | 3 | 0 | 3 |
| NKX2-3 | FXYD2 | 0.674091 | 4 | 0 | 4 |
| NKX2-3 | GALNT9 | 0.672694 | 3 | 0 | 3 |
| NKX2-3 | ZSWIM3 | 0.661394 | 3 | 0 | 3 |
| NKX2-3 | KCNA7 | 0.659926 | 4 | 0 | 4 |
For details and further investigation, click here