| Full name: nemo like kinase | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 17q11.2 | ||
| Entrez ID: 51701 | HGNC ID: HGNC:29858 | Ensembl Gene: ENSG00000087095 | OMIM ID: 609476 |
| Drug and gene relationship at DGIdb | |||
NLK involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04010 | MAPK signaling pathway | |
| hsa04068 | FoxO signaling pathway | |
| hsa04310 | Wnt signaling pathway | |
| hsa04520 | Adherens junction |
Expression of NLK:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NLK | 51701 | 222589_at | 0.4374 | 0.2213 | |
| GSE20347 | NLK | 51701 | 218318_s_at | 0.3295 | 0.0246 | |
| GSE23400 | NLK | 51701 | 218318_s_at | 0.1659 | 0.0000 | |
| GSE26886 | NLK | 51701 | 222589_at | 0.7653 | 0.0190 | |
| GSE29001 | NLK | 51701 | 218318_s_at | 0.4837 | 0.0345 | |
| GSE38129 | NLK | 51701 | 218318_s_at | 0.5648 | 0.0286 | |
| GSE45670 | NLK | 51701 | 222589_at | 0.3592 | 0.0315 | |
| GSE53622 | NLK | 51701 | 12813 | 0.2036 | 0.0007 | |
| GSE53624 | NLK | 51701 | 12813 | 0.2298 | 0.0000 | |
| GSE63941 | NLK | 51701 | 222589_at | -0.5556 | 0.3418 | |
| GSE77861 | NLK | 51701 | 222589_at | 0.6373 | 0.0495 | |
| GSE97050 | NLK | 51701 | A_33_P3327108 | 0.3525 | 0.1816 | |
| SRP007169 | NLK | 51701 | RNAseq | 0.2157 | 0.6403 | |
| SRP008496 | NLK | 51701 | RNAseq | 0.1622 | 0.5483 | |
| SRP064894 | NLK | 51701 | RNAseq | 0.6852 | 0.0000 | |
| SRP133303 | NLK | 51701 | RNAseq | 0.2934 | 0.0064 | |
| SRP159526 | NLK | 51701 | RNAseq | 0.5108 | 0.0081 | |
| SRP193095 | NLK | 51701 | RNAseq | 0.1585 | 0.0858 | |
| SRP219564 | NLK | 51701 | RNAseq | 0.3703 | 0.0788 | |
| TCGA | NLK | 51701 | RNAseq | -0.0817 | 0.1451 |
Upregulated datasets: 0; Downregulated datasets: 0.
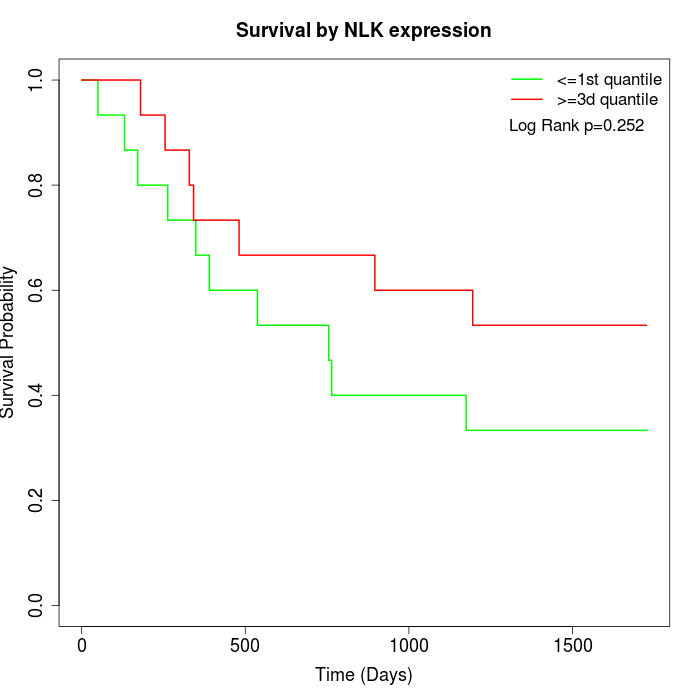
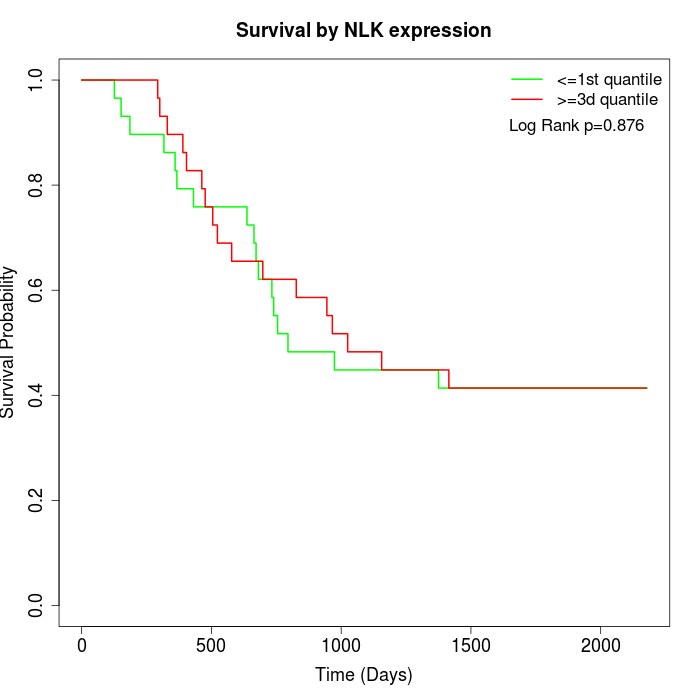
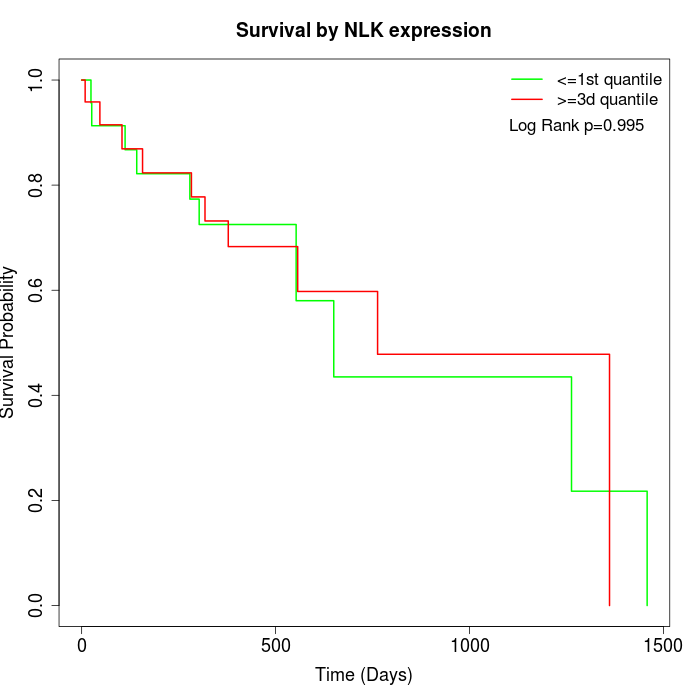
Survival by NLK expression:
Note: Click image to view full size file.
Copy number change of NLK:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NLK | 51701 | 9 | 1 | 20 | |
| GSE20123 | NLK | 51701 | 9 | 1 | 20 | |
| GSE43470 | NLK | 51701 | 0 | 1 | 42 | |
| GSE46452 | NLK | 51701 | 33 | 1 | 25 | |
| GSE47630 | NLK | 51701 | 7 | 1 | 32 | |
| GSE54993 | NLK | 51701 | 2 | 3 | 65 | |
| GSE54994 | NLK | 51701 | 7 | 6 | 40 | |
| GSE60625 | NLK | 51701 | 4 | 0 | 7 | |
| GSE74703 | NLK | 51701 | 0 | 1 | 35 | |
| GSE74704 | NLK | 51701 | 7 | 1 | 12 | |
| TCGA | NLK | 51701 | 21 | 10 | 65 |
Total number of gains: 99; Total number of losses: 26; Total Number of normals: 363.
Somatic mutations of NLK:
Generating mutation plots.
Highly correlated genes for NLK:
Showing top 20/1135 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NLK | POLR2A | 0.854659 | 3 | 0 | 3 |
| NLK | XAB2 | 0.852485 | 3 | 0 | 3 |
| NLK | SESTD1 | 0.817158 | 3 | 0 | 3 |
| NLK | ZNF766 | 0.799943 | 3 | 0 | 3 |
| NLK | GLO1 | 0.789963 | 3 | 0 | 3 |
| NLK | WDR25 | 0.789633 | 3 | 0 | 3 |
| NLK | NAF1 | 0.773634 | 3 | 0 | 3 |
| NLK | PRKCSH | 0.766457 | 3 | 0 | 3 |
| NLK | TGFBRAP1 | 0.765264 | 3 | 0 | 3 |
| NLK | CSNK1D | 0.763892 | 3 | 0 | 3 |
| NLK | GTPBP6 | 0.756323 | 3 | 0 | 3 |
| NLK | DAZAP2 | 0.748428 | 3 | 0 | 3 |
| NLK | EXOC6 | 0.746697 | 4 | 0 | 3 |
| NLK | RICTOR | 0.746373 | 3 | 0 | 3 |
| NLK | ORMDL3 | 0.739983 | 3 | 0 | 3 |
| NLK | SLC39A1 | 0.73881 | 3 | 0 | 3 |
| NLK | GMCL1 | 0.738554 | 3 | 0 | 3 |
| NLK | ZNF260 | 0.738358 | 5 | 0 | 5 |
| NLK | GDAP1 | 0.73231 | 3 | 0 | 3 |
| NLK | EIF2AK4 | 0.727355 | 3 | 0 | 3 |
For details and further investigation, click here