| Full name: NLR family pyrin domain containing 9 | Alias Symbol: NOD6|PAN12|CLR19.1 | ||
| Type: protein-coding gene | Cytoband: 19q13.43 | ||
| Entrez ID: 338321 | HGNC ID: HGNC:22941 | Ensembl Gene: ENSG00000185792 | OMIM ID: 609663 |
| Drug and gene relationship at DGIdb | |||
Expression of NLRP9:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NLRP9 | 338321 | 1553527_at | 0.0029 | 0.9931 | |
| GSE26886 | NLRP9 | 338321 | 1553527_at | 0.1684 | 0.1956 | |
| GSE45670 | NLRP9 | 338321 | 1553527_at | 0.0612 | 0.5988 | |
| GSE53622 | NLRP9 | 338321 | 44285 | 0.3589 | 0.0511 | |
| GSE53624 | NLRP9 | 338321 | 44285 | 0.2114 | 0.0838 | |
| GSE63941 | NLRP9 | 338321 | 1553527_at | 0.0300 | 0.8671 | |
| GSE77861 | NLRP9 | 338321 | 1553527_at | -0.2035 | 0.0888 | |
| TCGA | NLRP9 | 338321 | RNAseq | -2.1776 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 1.
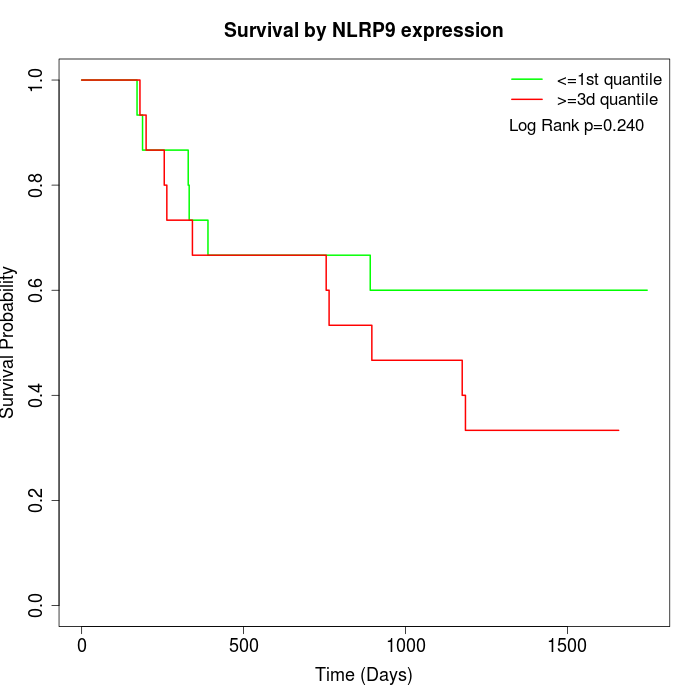
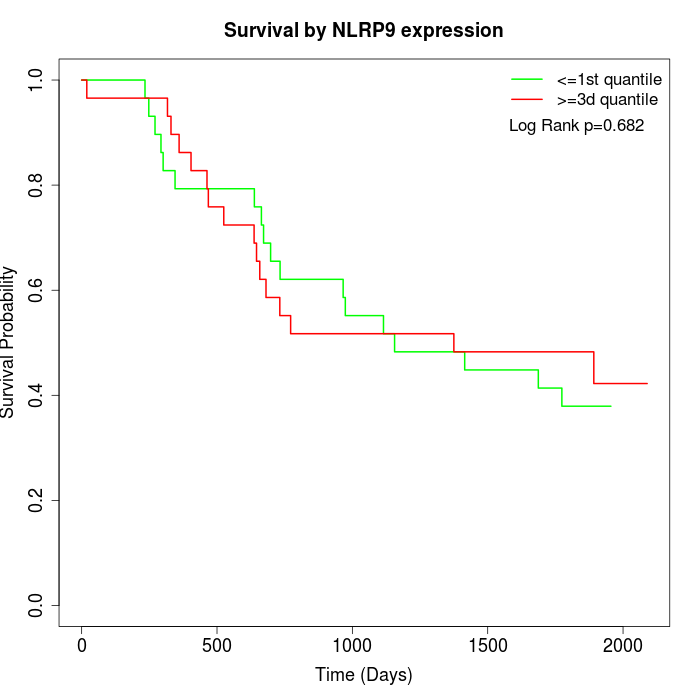
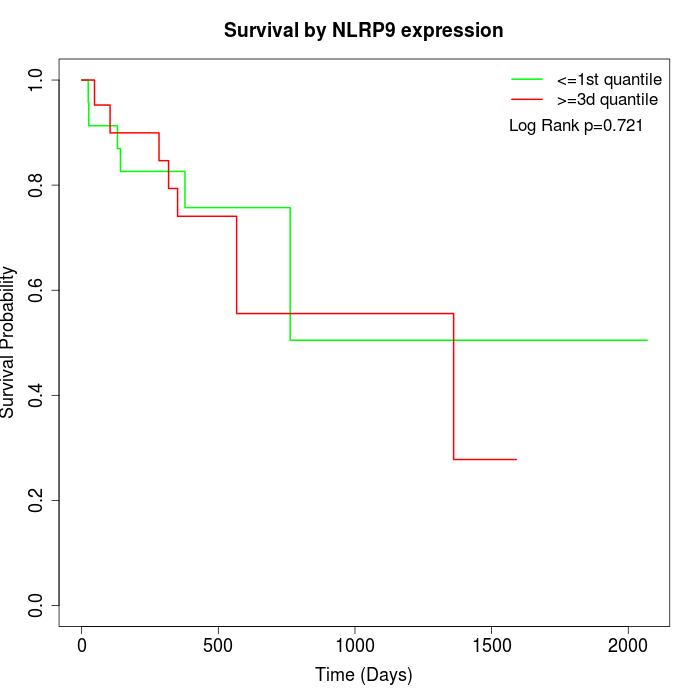
Survival by NLRP9 expression:
Note: Click image to view full size file.
Copy number change of NLRP9:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NLRP9 | 338321 | 3 | 4 | 23 | |
| GSE20123 | NLRP9 | 338321 | 3 | 3 | 24 | |
| GSE43470 | NLRP9 | 338321 | 3 | 9 | 31 | |
| GSE46452 | NLRP9 | 338321 | 45 | 1 | 13 | |
| GSE47630 | NLRP9 | 338321 | 8 | 6 | 26 | |
| GSE54993 | NLRP9 | 338321 | 17 | 4 | 49 | |
| GSE54994 | NLRP9 | 338321 | 4 | 13 | 36 | |
| GSE60625 | NLRP9 | 338321 | 9 | 0 | 2 | |
| GSE74703 | NLRP9 | 338321 | 3 | 6 | 27 | |
| GSE74704 | NLRP9 | 338321 | 3 | 1 | 16 | |
| TCGA | NLRP9 | 338321 | 19 | 15 | 62 |
Total number of gains: 117; Total number of losses: 62; Total Number of normals: 309.
Somatic mutations of NLRP9:
Generating mutation plots.
Highly correlated genes for NLRP9:
Showing top 20/54 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NLRP9 | TAS2R41 | 0.684431 | 3 | 0 | 3 |
| NLRP9 | C8orf74 | 0.667201 | 3 | 0 | 3 |
| NLRP9 | CRYGD | 0.656807 | 3 | 0 | 3 |
| NLRP9 | ALPI | 0.654415 | 3 | 0 | 3 |
| NLRP9 | FCN3 | 0.645188 | 3 | 0 | 3 |
| NLRP9 | OPN4 | 0.640258 | 5 | 0 | 4 |
| NLRP9 | TMEM105 | 0.639455 | 4 | 0 | 4 |
| NLRP9 | LINC00870 | 0.620889 | 3 | 0 | 3 |
| NLRP9 | LINC00421 | 0.619355 | 4 | 0 | 4 |
| NLRP9 | FAM181A-AS1 | 0.617081 | 4 | 0 | 3 |
| NLRP9 | BEND4 | 0.615476 | 3 | 0 | 3 |
| NLRP9 | RAPGEF3 | 0.610644 | 4 | 0 | 3 |
| NLRP9 | PCSK4 | 0.609774 | 4 | 0 | 3 |
| NLRP9 | LINC01107 | 0.599353 | 3 | 0 | 3 |
| NLRP9 | KRTAP2-1 | 0.594976 | 4 | 0 | 4 |
| NLRP9 | MYL3 | 0.593645 | 4 | 0 | 4 |
| NLRP9 | BRINP3 | 0.588927 | 3 | 0 | 3 |
| NLRP9 | LINC00856 | 0.588841 | 3 | 0 | 3 |
| NLRP9 | GBX2 | 0.587217 | 3 | 0 | 3 |
| NLRP9 | LINC01134 | 0.575108 | 4 | 0 | 3 |
For details and further investigation, click here