| Full name: nodal growth differentiation factor | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 10q22.1 | ||
| Entrez ID: 4838 | HGNC ID: HGNC:7865 | Ensembl Gene: ENSG00000156574 | OMIM ID: 601265 |
| Drug and gene relationship at DGIdb | |||
NODAL involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04350 | TGF-beta signaling pathway | |
| hsa04550 | Signaling pathways regulating pluripotency of stem cells |
Expression of NODAL:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NODAL | 4838 | 230916_at | -0.1003 | 0.6166 | |
| GSE26886 | NODAL | 4838 | 230916_at | -0.1741 | 0.1236 | |
| GSE45670 | NODAL | 4838 | 230916_at | 0.0139 | 0.9020 | |
| GSE53622 | NODAL | 4838 | 74366 | -0.3887 | 0.0000 | |
| GSE53624 | NODAL | 4838 | 74366 | -0.2953 | 0.0015 | |
| GSE63941 | NODAL | 4838 | 230916_at | 0.2608 | 0.1106 | |
| GSE77861 | NODAL | 4838 | 230916_at | -0.1544 | 0.2677 | |
| GSE97050 | NODAL | 4838 | A_33_P3299776 | 0.0651 | 0.7669 | |
| SRP133303 | NODAL | 4838 | RNAseq | -0.2064 | 0.4268 | |
| TCGA | NODAL | 4838 | RNAseq | 0.4704 | 0.1178 |
Upregulated datasets: 0; Downregulated datasets: 0.
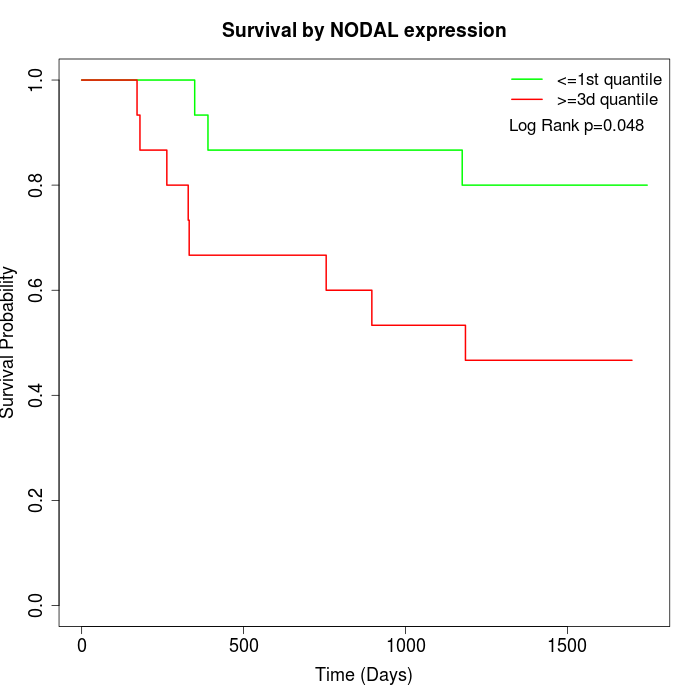
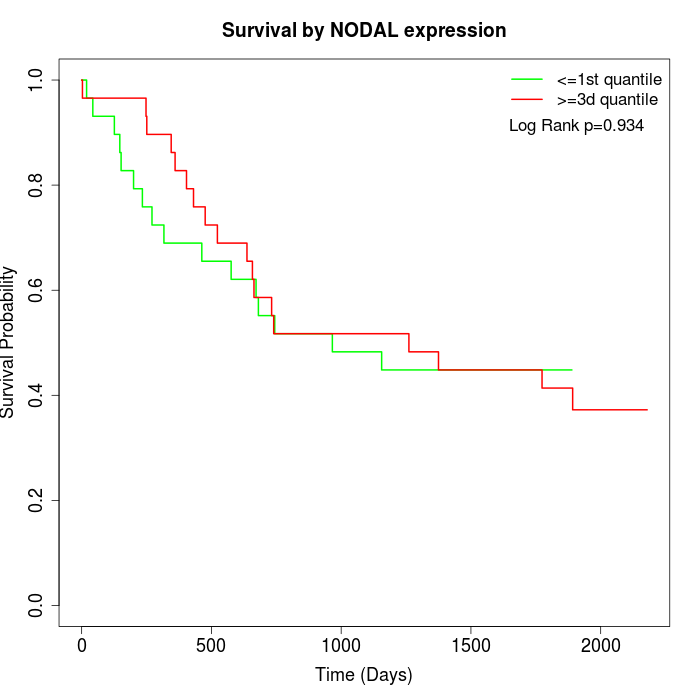
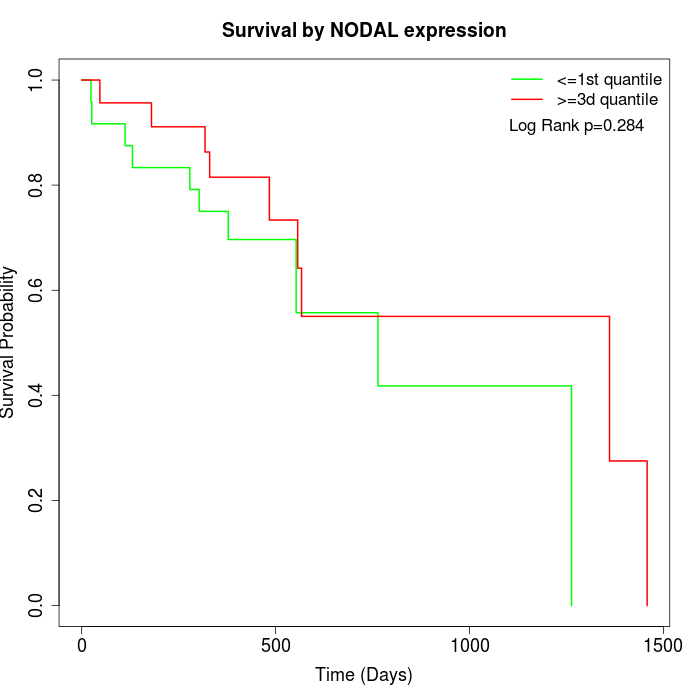
Survival by NODAL expression:
Note: Click image to view full size file.
Copy number change of NODAL:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NODAL | 4838 | 2 | 4 | 24 | |
| GSE20123 | NODAL | 4838 | 2 | 3 | 25 | |
| GSE43470 | NODAL | 4838 | 1 | 7 | 35 | |
| GSE46452 | NODAL | 4838 | 0 | 11 | 48 | |
| GSE47630 | NODAL | 4838 | 2 | 14 | 24 | |
| GSE54993 | NODAL | 4838 | 7 | 0 | 63 | |
| GSE54994 | NODAL | 4838 | 2 | 12 | 39 | |
| GSE60625 | NODAL | 4838 | 0 | 0 | 11 | |
| GSE74703 | NODAL | 4838 | 1 | 4 | 31 | |
| GSE74704 | NODAL | 4838 | 1 | 1 | 18 | |
| TCGA | NODAL | 4838 | 9 | 23 | 64 |
Total number of gains: 27; Total number of losses: 79; Total Number of normals: 382.
Somatic mutations of NODAL:
Generating mutation plots.
Highly correlated genes for NODAL:
Showing top 20/161 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NODAL | LRRC29 | 0.659173 | 4 | 0 | 3 |
| NODAL | FASLG | 0.657795 | 3 | 0 | 3 |
| NODAL | LINC01214 | 0.650087 | 3 | 0 | 3 |
| NODAL | FOXB1 | 0.647892 | 3 | 0 | 3 |
| NODAL | C11orf21 | 0.647321 | 4 | 0 | 3 |
| NODAL | LSMEM2 | 0.644446 | 3 | 0 | 3 |
| NODAL | RAX | 0.643926 | 4 | 0 | 3 |
| NODAL | LELP1 | 0.631548 | 3 | 0 | 3 |
| NODAL | MMP17 | 0.628541 | 3 | 0 | 3 |
| NODAL | GBX2 | 0.625871 | 3 | 0 | 3 |
| NODAL | CD300LG | 0.625271 | 4 | 0 | 4 |
| NODAL | G6PC | 0.616151 | 3 | 0 | 3 |
| NODAL | IQCD | 0.615821 | 4 | 0 | 3 |
| NODAL | CDH10 | 0.603769 | 3 | 0 | 3 |
| NODAL | TEX13B | 0.6029 | 3 | 0 | 3 |
| NODAL | PTPRN | 0.602408 | 4 | 0 | 3 |
| NODAL | PHGR1 | 0.594535 | 4 | 0 | 3 |
| NODAL | CHRNA2 | 0.594266 | 6 | 0 | 5 |
| NODAL | DSCAM | 0.594234 | 4 | 0 | 4 |
| NODAL | DNAI1 | 0.593241 | 3 | 0 | 3 |
For details and further investigation, click here