| Full name: late cornified envelope like proline rich 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 1q21.3 | ||
| Entrez ID: 149018 | HGNC ID: HGNC:32046 | Ensembl Gene: ENSG00000203784 | OMIM ID: 611042 |
| Drug and gene relationship at DGIdb | |||
Expression of LELP1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LELP1 | 149018 | 231363_at | -0.1957 | 0.4188 | |
| GSE26886 | LELP1 | 149018 | 231363_at | 0.1406 | 0.2153 | |
| GSE45670 | LELP1 | 149018 | 231363_at | -0.0637 | 0.5470 | |
| GSE53622 | LELP1 | 149018 | 17875 | -0.0223 | 0.8454 | |
| GSE53624 | LELP1 | 149018 | 17875 | -0.0632 | 0.7050 | |
| GSE63941 | LELP1 | 149018 | 231363_at | 0.1470 | 0.4021 | |
| GSE77861 | LELP1 | 149018 | 231363_at | -0.1969 | 0.0514 | |
| GSE97050 | LELP1 | 149018 | A_23_P51311 | 0.0501 | 0.8599 |
Upregulated datasets: 0; Downregulated datasets: 0.
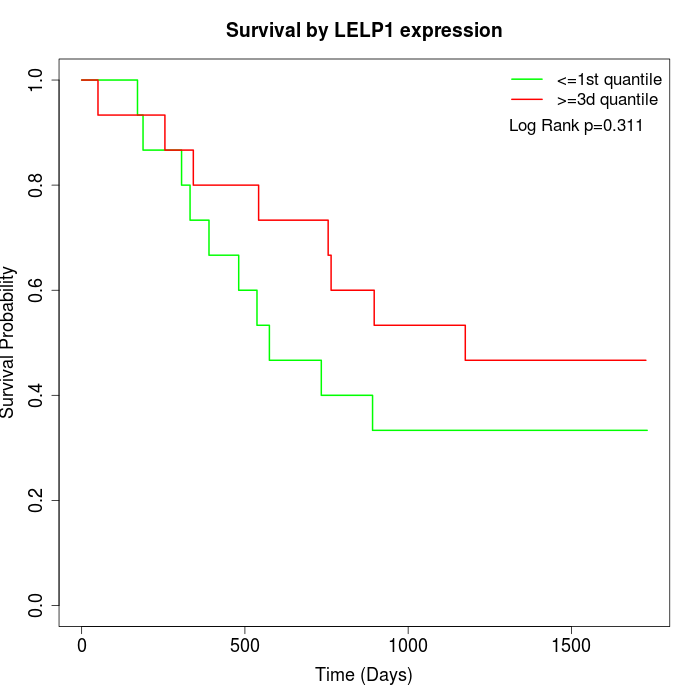
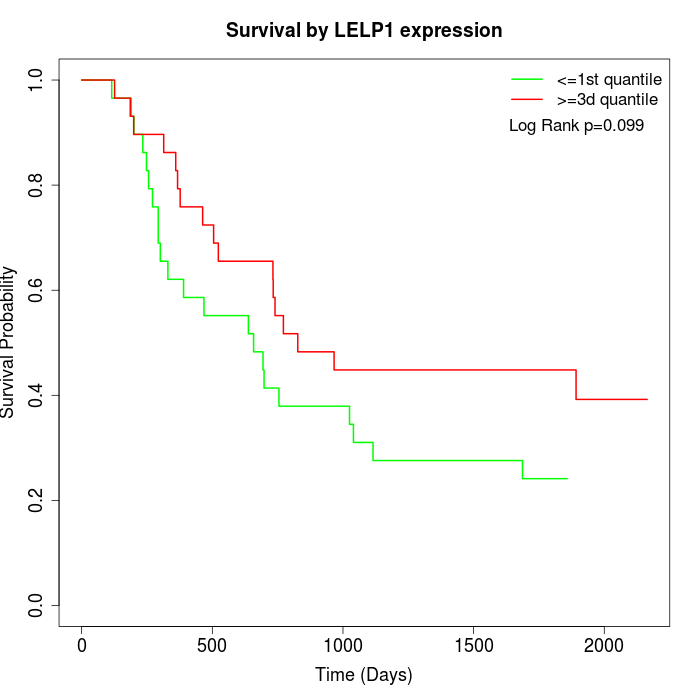
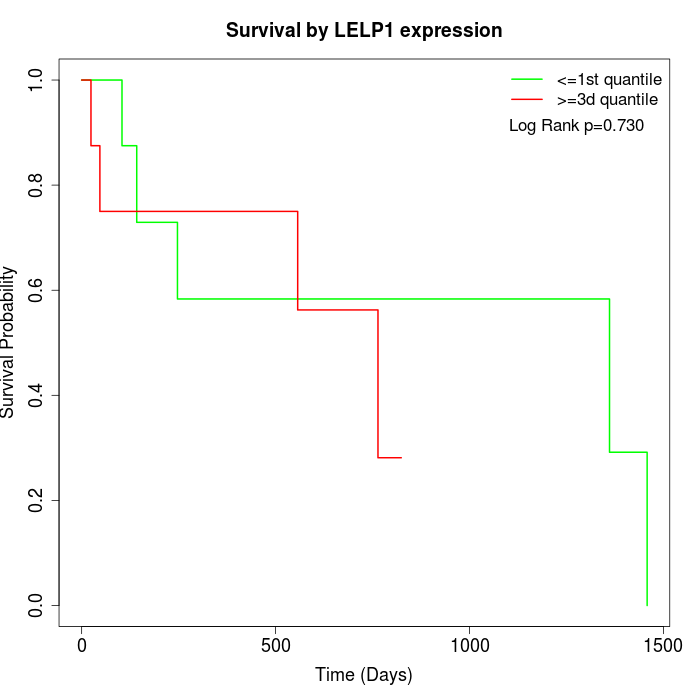
Survival by LELP1 expression:
Note: Click image to view full size file.
Copy number change of LELP1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LELP1 | 149018 | 15 | 0 | 15 | |
| GSE20123 | LELP1 | 149018 | 15 | 0 | 15 | |
| GSE43470 | LELP1 | 149018 | 6 | 2 | 35 | |
| GSE46452 | LELP1 | 149018 | 2 | 1 | 56 | |
| GSE47630 | LELP1 | 149018 | 15 | 0 | 25 | |
| GSE54993 | LELP1 | 149018 | 0 | 4 | 66 | |
| GSE54994 | LELP1 | 149018 | 16 | 0 | 37 | |
| GSE60625 | LELP1 | 149018 | 0 | 0 | 11 | |
| GSE74703 | LELP1 | 149018 | 6 | 1 | 29 | |
| GSE74704 | LELP1 | 149018 | 7 | 0 | 13 | |
| TCGA | LELP1 | 149018 | 39 | 2 | 55 |
Total number of gains: 121; Total number of losses: 10; Total Number of normals: 357.
Somatic mutations of LELP1:
Generating mutation plots.
Highly correlated genes for LELP1:
Showing top 20/315 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LELP1 | PHF7 | 0.836371 | 3 | 0 | 3 |
| LELP1 | CTSE | 0.826027 | 3 | 0 | 3 |
| LELP1 | MYO1A | 0.816479 | 3 | 0 | 3 |
| LELP1 | POU4F3 | 0.787492 | 3 | 0 | 3 |
| LELP1 | TULP2 | 0.76906 | 3 | 0 | 3 |
| LELP1 | NTRK1 | 0.76228 | 3 | 0 | 3 |
| LELP1 | SCARF2 | 0.759778 | 3 | 0 | 3 |
| LELP1 | ABCC8 | 0.756125 | 4 | 0 | 4 |
| LELP1 | VEPH1 | 0.743354 | 3 | 0 | 3 |
| LELP1 | DCD | 0.740272 | 3 | 0 | 3 |
| LELP1 | SCN2B | 0.739269 | 4 | 0 | 3 |
| LELP1 | TGM6 | 0.736009 | 4 | 0 | 4 |
| LELP1 | ZGLP1 | 0.730438 | 4 | 0 | 3 |
| LELP1 | PCDH1 | 0.727812 | 4 | 0 | 4 |
| LELP1 | TEKT2 | 0.713371 | 3 | 0 | 3 |
| LELP1 | HAAO | 0.711992 | 4 | 0 | 3 |
| LELP1 | CABP7 | 0.71095 | 3 | 0 | 3 |
| LELP1 | LINC00494 | 0.709419 | 3 | 0 | 3 |
| LELP1 | LMAN1L | 0.707704 | 3 | 0 | 3 |
| LELP1 | CPA5 | 0.695317 | 3 | 0 | 3 |
For details and further investigation, click here