| Full name: neuronal PAS domain protein 4 | Alias Symbol: PASD10|NXF|Le-PAS|bHLHe79 | ||
| Type: protein-coding gene | Cytoband: 11q13.2 | ||
| Entrez ID: 266743 | HGNC ID: HGNC:18983 | Ensembl Gene: ENSG00000174576 | OMIM ID: 608554 |
| Drug and gene relationship at DGIdb | |||
Expression of NPAS4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NPAS4 | 266743 | 240794_at | 0.0664 | 0.7288 | |
| GSE26886 | NPAS4 | 266743 | 240794_at | -0.0260 | 0.8499 | |
| GSE45670 | NPAS4 | 266743 | 240794_at | 0.1458 | 0.1406 | |
| GSE53622 | NPAS4 | 266743 | 36493 | -0.2714 | 0.0086 | |
| GSE53624 | NPAS4 | 266743 | 36493 | -0.4019 | 0.0000 | |
| GSE63941 | NPAS4 | 266743 | 240794_at | 0.1155 | 0.5348 | |
| GSE77861 | NPAS4 | 266743 | 240794_at | -0.0687 | 0.7190 | |
| SRP159526 | NPAS4 | 266743 | RNAseq | -1.7203 | 0.0463 | |
| SRP219564 | NPAS4 | 266743 | RNAseq | -1.0618 | 0.3750 | |
| TCGA | NPAS4 | 266743 | RNAseq | -2.0788 | 0.0005 |
Upregulated datasets: 0; Downregulated datasets: 2.
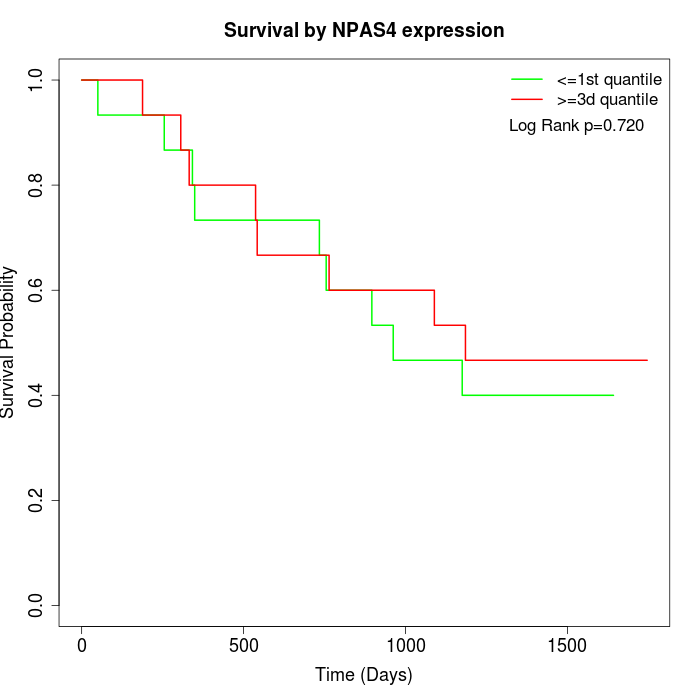
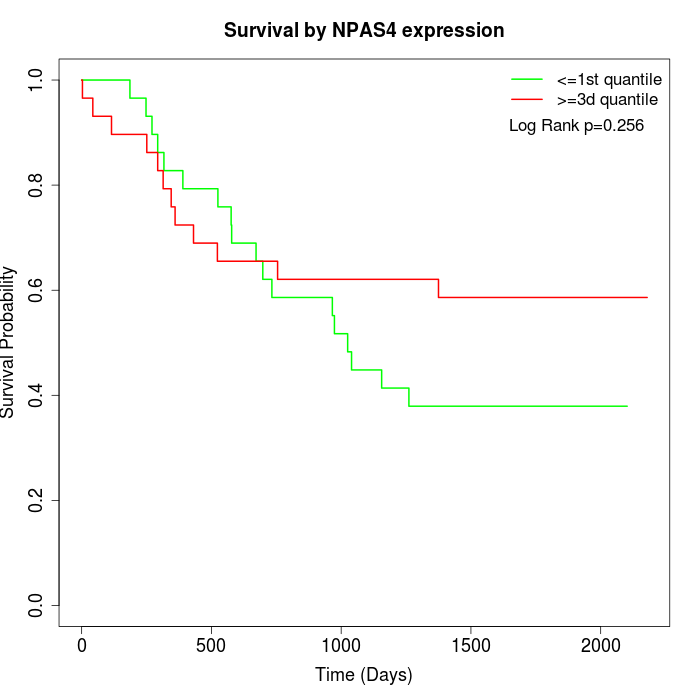
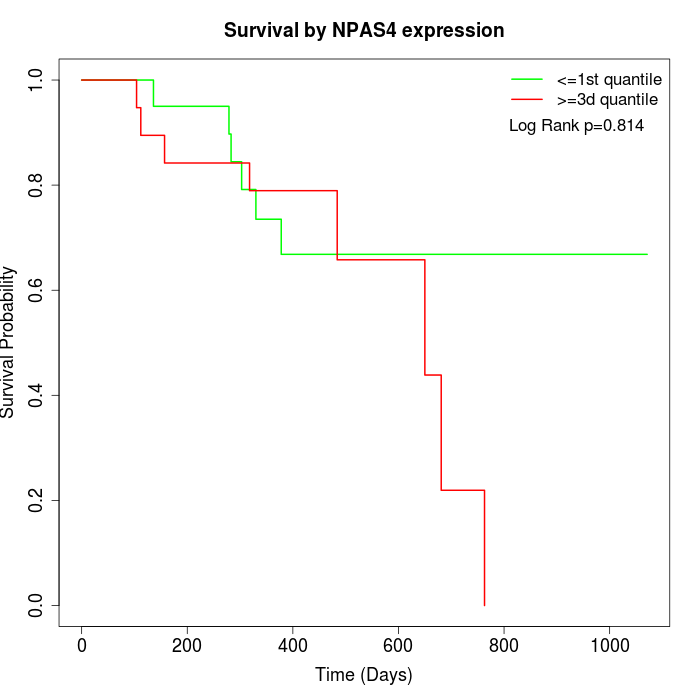
Survival by NPAS4 expression:
Note: Click image to view full size file.
Copy number change of NPAS4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NPAS4 | 266743 | 9 | 4 | 17 | |
| GSE20123 | NPAS4 | 266743 | 9 | 4 | 17 | |
| GSE43470 | NPAS4 | 266743 | 6 | 1 | 36 | |
| GSE46452 | NPAS4 | 266743 | 12 | 3 | 44 | |
| GSE47630 | NPAS4 | 266743 | 9 | 4 | 27 | |
| GSE54993 | NPAS4 | 266743 | 3 | 0 | 67 | |
| GSE54994 | NPAS4 | 266743 | 9 | 5 | 39 | |
| GSE60625 | NPAS4 | 266743 | 0 | 3 | 8 | |
| GSE74703 | NPAS4 | 266743 | 4 | 0 | 32 | |
| GSE74704 | NPAS4 | 266743 | 7 | 2 | 11 | |
| TCGA | NPAS4 | 266743 | 26 | 6 | 64 |
Total number of gains: 94; Total number of losses: 32; Total Number of normals: 362.
Somatic mutations of NPAS4:
Generating mutation plots.
Highly correlated genes for NPAS4:
Showing top 20/67 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NPAS4 | GPR26 | 0.664466 | 3 | 0 | 3 |
| NPAS4 | UPB1 | 0.661972 | 3 | 0 | 3 |
| NPAS4 | CELF3 | 0.655105 | 3 | 0 | 3 |
| NPAS4 | KLK2 | 0.653276 | 3 | 0 | 3 |
| NPAS4 | DKKL1 | 0.652143 | 3 | 0 | 3 |
| NPAS4 | FAM153A | 0.650601 | 4 | 0 | 4 |
| NPAS4 | ICAM5 | 0.644231 | 3 | 0 | 3 |
| NPAS4 | FGF18 | 0.639468 | 3 | 0 | 3 |
| NPAS4 | MASP1 | 0.638163 | 3 | 0 | 3 |
| NPAS4 | KRT84 | 0.632012 | 3 | 0 | 3 |
| NPAS4 | FIBCD1 | 0.63018 | 3 | 0 | 3 |
| NPAS4 | ZBTB7B | 0.624511 | 3 | 0 | 3 |
| NPAS4 | MYL10 | 0.622228 | 3 | 0 | 3 |
| NPAS4 | MMP24 | 0.620785 | 3 | 0 | 3 |
| NPAS4 | SNORA71A | 0.619272 | 3 | 0 | 3 |
| NPAS4 | SCUBE1 | 0.617613 | 3 | 0 | 3 |
| NPAS4 | FAM171A2 | 0.598063 | 3 | 0 | 3 |
| NPAS4 | ANKRD53 | 0.597492 | 3 | 0 | 3 |
| NPAS4 | RIMS4 | 0.595549 | 4 | 0 | 3 |
| NPAS4 | MUC8 | 0.594653 | 3 | 0 | 3 |
For details and further investigation, click here