| Full name: NUT midline carcinoma family member 1 | Alias Symbol: NUT|DKFZp434O192|FAM22H | ||
| Type: protein-coding gene | Cytoband: 15q14 | ||
| Entrez ID: 256646 | HGNC ID: HGNC:29919 | Ensembl Gene: ENSG00000184507 | OMIM ID: 608963 |
| Drug and gene relationship at DGIdb | |||
Expression of NUTM1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NUTM1 | 256646 | 231338_at | -0.0746 | 0.7655 | |
| GSE26886 | NUTM1 | 256646 | 231338_at | 0.2204 | 0.0697 | |
| GSE45670 | NUTM1 | 256646 | 1564603_at | 0.1571 | 0.1683 | |
| GSE53622 | NUTM1 | 256646 | 30185 | 0.2034 | 0.0249 | |
| GSE53624 | NUTM1 | 256646 | 30185 | 0.0593 | 0.5511 | |
| GSE63941 | NUTM1 | 256646 | 231338_at | 0.3548 | 0.0268 | |
| GSE77861 | NUTM1 | 256646 | 1564603_at | -0.1204 | 0.4452 |
Upregulated datasets: 0; Downregulated datasets: 0.
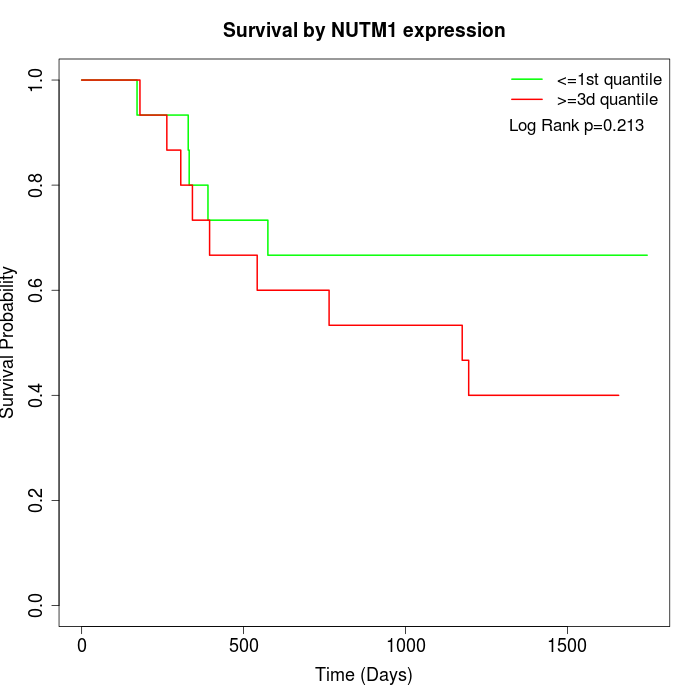
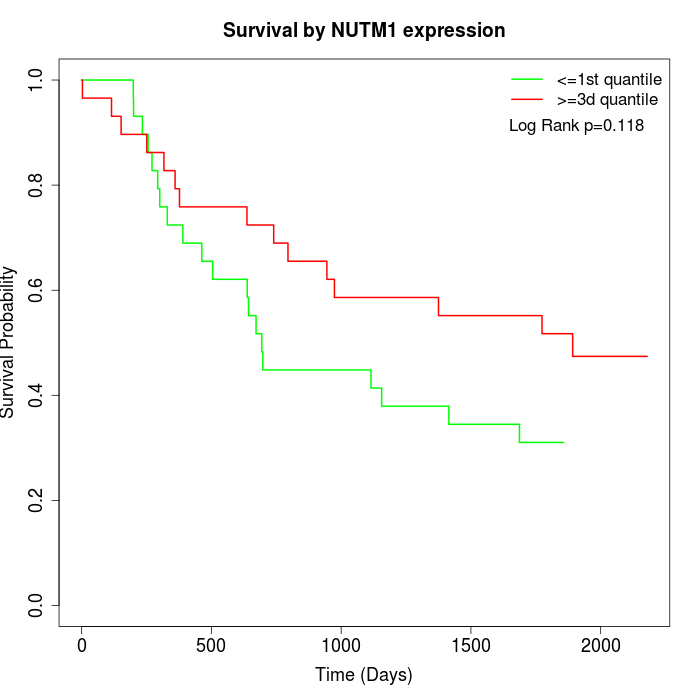
Survival by NUTM1 expression:
Note: Click image to view full size file.
Copy number change of NUTM1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NUTM1 | 256646 | 3 | 6 | 21 | |
| GSE20123 | NUTM1 | 256646 | 3 | 6 | 21 | |
| GSE43470 | NUTM1 | 256646 | 4 | 4 | 35 | |
| GSE46452 | NUTM1 | 256646 | 3 | 7 | 49 | |
| GSE47630 | NUTM1 | 256646 | 8 | 11 | 21 | |
| GSE54993 | NUTM1 | 256646 | 4 | 6 | 60 | |
| GSE54994 | NUTM1 | 256646 | 5 | 8 | 40 | |
| GSE60625 | NUTM1 | 256646 | 4 | 0 | 7 | |
| GSE74703 | NUTM1 | 256646 | 4 | 3 | 29 | |
| GSE74704 | NUTM1 | 256646 | 2 | 5 | 13 | |
| TCGA | NUTM1 | 256646 | 13 | 17 | 66 |
Total number of gains: 53; Total number of losses: 73; Total Number of normals: 362.
Somatic mutations of NUTM1:
Generating mutation plots.
Highly correlated genes for NUTM1:
Showing top 20/499 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NUTM1 | GPR182 | 0.778845 | 4 | 0 | 4 |
| NUTM1 | DSCR10 | 0.7725 | 4 | 0 | 4 |
| NUTM1 | CPLX2 | 0.749882 | 3 | 0 | 3 |
| NUTM1 | ZAN | 0.746824 | 3 | 0 | 3 |
| NUTM1 | LINC00486 | 0.730789 | 3 | 0 | 3 |
| NUTM1 | BARHL1 | 0.723246 | 4 | 0 | 4 |
| NUTM1 | LINC00523 | 0.719795 | 5 | 0 | 4 |
| NUTM1 | CASS4 | 0.716254 | 3 | 0 | 3 |
| NUTM1 | CAMKV | 0.707331 | 4 | 0 | 4 |
| NUTM1 | TSPAN11 | 0.704199 | 4 | 0 | 4 |
| NUTM1 | F2RL3 | 0.701291 | 4 | 0 | 4 |
| NUTM1 | THRA | 0.70113 | 4 | 0 | 3 |
| NUTM1 | KRTAP4-8 | 0.700085 | 5 | 0 | 4 |
| NUTM1 | GLP1R | 0.689121 | 3 | 0 | 3 |
| NUTM1 | CLDN9 | 0.688711 | 4 | 0 | 3 |
| NUTM1 | PGP | 0.686951 | 3 | 0 | 3 |
| NUTM1 | KCNH3 | 0.683392 | 5 | 0 | 4 |
| NUTM1 | CECR9 | 0.681997 | 4 | 0 | 3 |
| NUTM1 | TTC7A | 0.68045 | 3 | 0 | 3 |
| NUTM1 | TNNI1 | 0.680377 | 4 | 0 | 4 |
For details and further investigation, click here