| Full name: thyroid hormone receptor alpha | Alias Symbol: EAR-7.1/EAR-7.2|THRA3|AR7|ERBA|NR1A1 | ||
| Type: protein-coding gene | Cytoband: 17q21.1 | ||
| Entrez ID: 7067 | HGNC ID: HGNC:11796 | Ensembl Gene: ENSG00000126351 | OMIM ID: 190120 |
| Drug and gene relationship at DGIdb | |||
THRA involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04919 | Thyroid hormone signaling pathway |
Expression of THRA:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | THRA | 7067 | 35846_at | -0.3695 | 0.5062 | |
| GSE20347 | THRA | 7067 | 35846_at | 0.0685 | 0.5741 | |
| GSE23400 | THRA | 7067 | 214883_at | -0.1420 | 0.0000 | |
| GSE26886 | THRA | 7067 | 35846_at | 0.3934 | 0.0050 | |
| GSE29001 | THRA | 7067 | 214883_at | -0.2215 | 0.1470 | |
| GSE38129 | THRA | 7067 | 35846_at | -0.3048 | 0.0126 | |
| GSE45670 | THRA | 7067 | 35846_at | -0.3422 | 0.0008 | |
| GSE53622 | THRA | 7067 | 34811 | 0.1656 | 0.0241 | |
| GSE53624 | THRA | 7067 | 34811 | -0.1474 | 0.0479 | |
| GSE63941 | THRA | 7067 | 35846_at | -0.8015 | 0.0023 | |
| GSE77861 | THRA | 7067 | 1316_at | 0.0405 | 0.7271 | |
| GSE97050 | THRA | 7067 | A_24_P262407 | -0.5174 | 0.2508 | |
| SRP007169 | THRA | 7067 | RNAseq | 0.1928 | 0.6929 | |
| SRP008496 | THRA | 7067 | RNAseq | 0.0074 | 0.9826 | |
| SRP064894 | THRA | 7067 | RNAseq | -0.2479 | 0.1411 | |
| SRP133303 | THRA | 7067 | RNAseq | -0.4016 | 0.0424 | |
| SRP159526 | THRA | 7067 | RNAseq | -0.3525 | 0.1338 | |
| SRP193095 | THRA | 7067 | RNAseq | 0.3155 | 0.1558 | |
| SRP219564 | THRA | 7067 | RNAseq | -0.4266 | 0.4600 | |
| TCGA | THRA | 7067 | RNAseq | -0.4451 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 0.
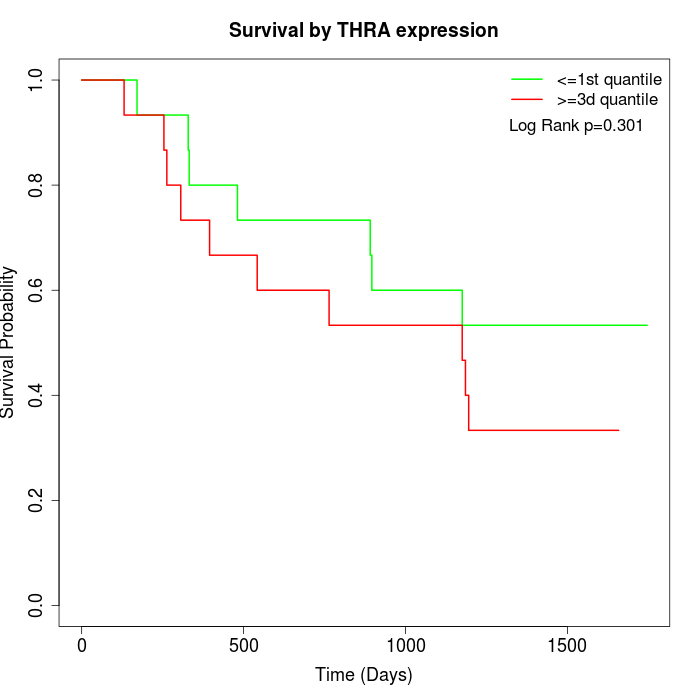
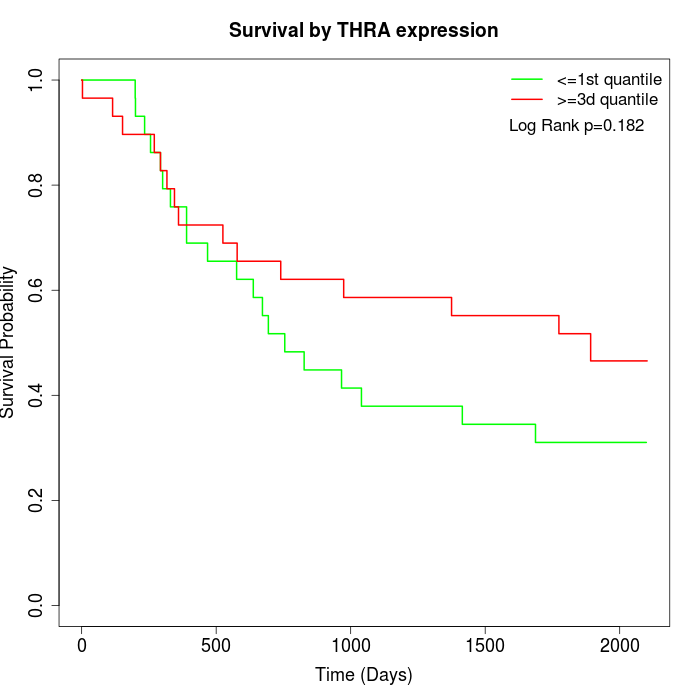
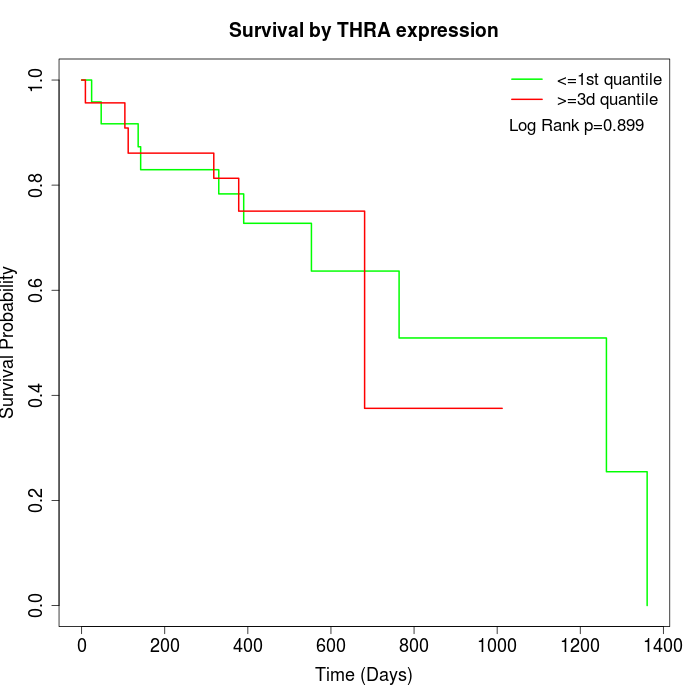
Survival by THRA expression:
Note: Click image to view full size file.
Copy number change of THRA:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | THRA | 7067 | 9 | 1 | 20 | |
| GSE20123 | THRA | 7067 | 9 | 1 | 20 | |
| GSE43470 | THRA | 7067 | 1 | 2 | 40 | |
| GSE46452 | THRA | 7067 | 34 | 0 | 25 | |
| GSE47630 | THRA | 7067 | 8 | 1 | 31 | |
| GSE54993 | THRA | 7067 | 3 | 4 | 63 | |
| GSE54994 | THRA | 7067 | 9 | 5 | 39 | |
| GSE60625 | THRA | 7067 | 4 | 0 | 7 | |
| GSE74703 | THRA | 7067 | 1 | 1 | 34 | |
| GSE74704 | THRA | 7067 | 5 | 1 | 14 | |
| TCGA | THRA | 7067 | 23 | 8 | 65 |
Total number of gains: 106; Total number of losses: 24; Total Number of normals: 358.
Somatic mutations of THRA:
Generating mutation plots.
Highly correlated genes for THRA:
Showing top 20/640 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| THRA | INSC | 0.834438 | 3 | 0 | 3 |
| THRA | PCA3 | 0.769887 | 3 | 0 | 3 |
| THRA | CPLX2 | 0.75451 | 3 | 0 | 3 |
| THRA | PARD3B | 0.750312 | 3 | 0 | 3 |
| THRA | C1QTNF7 | 0.745039 | 3 | 0 | 3 |
| THRA | SPRED3 | 0.736872 | 3 | 0 | 3 |
| THRA | CCDC136 | 0.736298 | 4 | 0 | 3 |
| THRA | NEGR1 | 0.725588 | 4 | 0 | 4 |
| THRA | HS1BP3 | 0.725176 | 5 | 0 | 5 |
| THRA | XKR4 | 0.717968 | 3 | 0 | 3 |
| THRA | PPP1R14A | 0.717914 | 4 | 0 | 3 |
| THRA | ABCA6 | 0.713515 | 6 | 0 | 6 |
| THRA | KRTAP10-9 | 0.711579 | 3 | 0 | 3 |
| THRA | ITPKB | 0.709968 | 3 | 0 | 3 |
| THRA | DUSP8 | 0.708415 | 3 | 0 | 3 |
| THRA | SH3GL2 | 0.707866 | 3 | 0 | 3 |
| THRA | ESRRG | 0.701321 | 4 | 0 | 3 |
| THRA | NUTM1 | 0.70113 | 4 | 0 | 3 |
| THRA | KCNN3 | 0.700142 | 4 | 0 | 3 |
| THRA | SNPH | 0.698913 | 6 | 0 | 5 |
For details and further investigation, click here