| Full name: purinergic receptor P2Y1 | Alias Symbol: P2Y1|SARCC | ||
| Type: protein-coding gene | Cytoband: 3q25.2 | ||
| Entrez ID: 5028 | HGNC ID: HGNC:8539 | Ensembl Gene: ENSG00000169860 | OMIM ID: 601167 |
| Drug and gene relationship at DGIdb | |||
P2RY1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04015 | Rap1 signaling pathway | |
| hsa04611 | Platelet activation | |
| hsa04742 | Taste transduction |
Expression of P2RY1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | P2RY1 | 5028 | 207455_at | 0.0390 | 0.9770 | |
| GSE20347 | P2RY1 | 5028 | 207455_at | -0.4030 | 0.0204 | |
| GSE23400 | P2RY1 | 5028 | 207455_at | -0.0459 | 0.4920 | |
| GSE26886 | P2RY1 | 5028 | 207455_at | -2.6167 | 0.0000 | |
| GSE29001 | P2RY1 | 5028 | 207455_at | -0.3963 | 0.4689 | |
| GSE38129 | P2RY1 | 5028 | 207455_at | -0.3062 | 0.1363 | |
| GSE45670 | P2RY1 | 5028 | 207455_at | 0.6616 | 0.0544 | |
| GSE53622 | P2RY1 | 5028 | 22340 | -0.7781 | 0.0007 | |
| GSE53624 | P2RY1 | 5028 | 22340 | -1.0374 | 0.0000 | |
| GSE63941 | P2RY1 | 5028 | 207455_at | 0.2978 | 0.5998 | |
| GSE77861 | P2RY1 | 5028 | 207455_at | 0.1082 | 0.7824 | |
| GSE97050 | P2RY1 | 5028 | A_23_P382835 | -0.2757 | 0.3718 | |
| SRP007169 | P2RY1 | 5028 | RNAseq | -0.1352 | 0.8202 | |
| SRP008496 | P2RY1 | 5028 | RNAseq | -0.4431 | 0.2061 | |
| SRP064894 | P2RY1 | 5028 | RNAseq | -1.1239 | 0.0062 | |
| SRP133303 | P2RY1 | 5028 | RNAseq | -0.5106 | 0.1353 | |
| SRP159526 | P2RY1 | 5028 | RNAseq | -0.9443 | 0.0724 | |
| SRP193095 | P2RY1 | 5028 | RNAseq | -0.7227 | 0.0842 | |
| SRP219564 | P2RY1 | 5028 | RNAseq | -1.3394 | 0.1166 | |
| TCGA | P2RY1 | 5028 | RNAseq | 0.8678 | 0.0005 |
Upregulated datasets: 0; Downregulated datasets: 3.
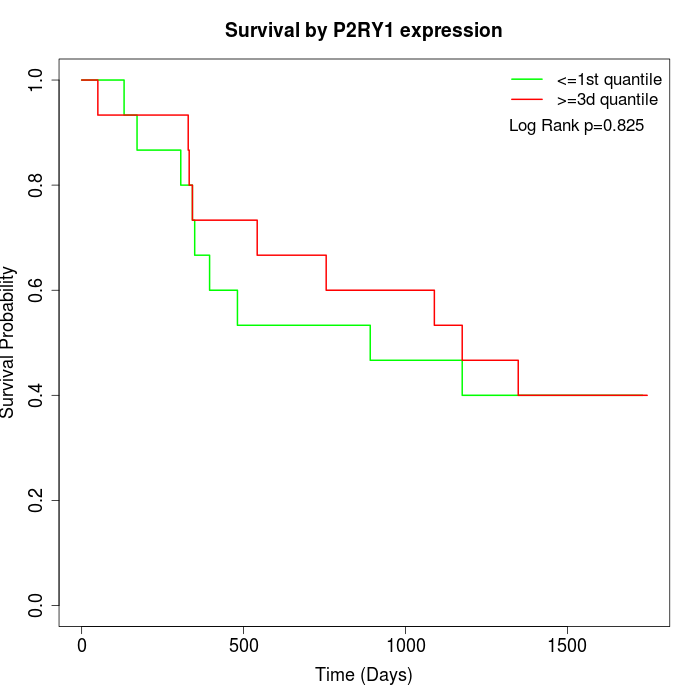
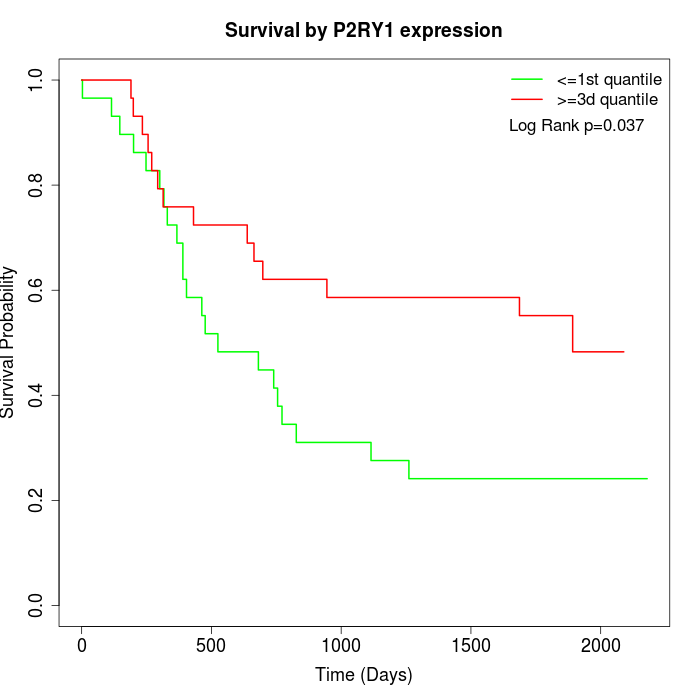
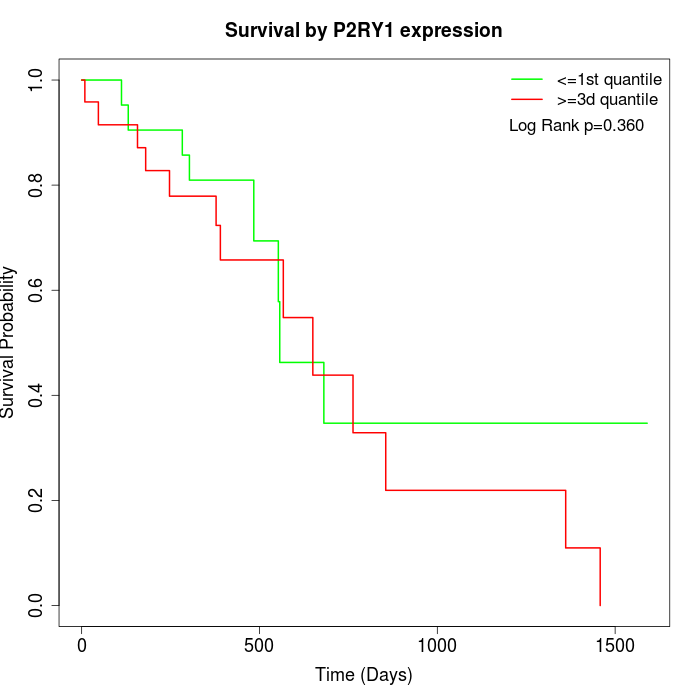
Survival by P2RY1 expression:
Note: Click image to view full size file.
Copy number change of P2RY1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | P2RY1 | 5028 | 21 | 0 | 9 | |
| GSE20123 | P2RY1 | 5028 | 21 | 0 | 9 | |
| GSE43470 | P2RY1 | 5028 | 24 | 0 | 19 | |
| GSE46452 | P2RY1 | 5028 | 18 | 2 | 39 | |
| GSE47630 | P2RY1 | 5028 | 19 | 3 | 18 | |
| GSE54993 | P2RY1 | 5028 | 1 | 10 | 59 | |
| GSE54994 | P2RY1 | 5028 | 38 | 1 | 14 | |
| GSE60625 | P2RY1 | 5028 | 0 | 6 | 5 | |
| GSE74703 | P2RY1 | 5028 | 21 | 0 | 15 | |
| GSE74704 | P2RY1 | 5028 | 13 | 0 | 7 | |
| TCGA | P2RY1 | 5028 | 72 | 1 | 23 |
Total number of gains: 248; Total number of losses: 23; Total Number of normals: 217.
Somatic mutations of P2RY1:
Generating mutation plots.
Highly correlated genes for P2RY1:
Showing top 20/525 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| P2RY1 | MAB21L3 | 0.753447 | 3 | 0 | 3 |
| P2RY1 | VSIG10L | 0.749851 | 4 | 0 | 4 |
| P2RY1 | TMEM79 | 0.742128 | 4 | 0 | 4 |
| P2RY1 | DEGS2 | 0.71672 | 4 | 0 | 4 |
| P2RY1 | TMPRSS11A | 0.715614 | 3 | 0 | 3 |
| P2RY1 | SPRR2D | 0.714033 | 3 | 0 | 3 |
| P2RY1 | SPRR2E | 0.713278 | 3 | 0 | 3 |
| P2RY1 | SPRR2A | 0.713216 | 3 | 0 | 3 |
| P2RY1 | SBSN | 0.706177 | 4 | 0 | 4 |
| P2RY1 | CYP2C9 | 0.700644 | 6 | 0 | 6 |
| P2RY1 | OTOP3 | 0.699471 | 3 | 0 | 3 |
| P2RY1 | SPRR2F | 0.699466 | 3 | 0 | 3 |
| P2RY1 | PRSS8 | 0.695928 | 3 | 0 | 3 |
| P2RY1 | NCCRP1 | 0.693717 | 3 | 0 | 3 |
| P2RY1 | RHBDL2 | 0.689114 | 5 | 0 | 5 |
| P2RY1 | PGM2 | 0.685492 | 3 | 0 | 3 |
| P2RY1 | FAM3D | 0.685245 | 4 | 0 | 4 |
| P2RY1 | CNFN | 0.683868 | 5 | 0 | 4 |
| P2RY1 | MUC15 | 0.681728 | 5 | 0 | 4 |
| P2RY1 | FLG-AS1 | 0.680727 | 3 | 0 | 3 |
For details and further investigation, click here