| Full name: proliferation and apoptosis adaptor protein 15 | Alias Symbol: HMAT1|MAT1|PED|PEA-15|MAT1H|HUMMAT1H|PED-PEA15|PED/PEA15 | ||
| Type: protein-coding gene | Cytoband: 1q23.2 | ||
| Entrez ID: 8682 | HGNC ID: HGNC:8822 | Ensembl Gene: ENSG00000162734 | OMIM ID: 603434 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of PEA15:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PEA15 | 8682 | 200788_s_at | -0.2347 | 0.3569 | |
| GSE20347 | PEA15 | 8682 | 200788_s_at | -0.5888 | 0.0000 | |
| GSE23400 | PEA15 | 8682 | 200788_s_at | -0.2121 | 0.0007 | |
| GSE26886 | PEA15 | 8682 | 200788_s_at | -1.1351 | 0.0000 | |
| GSE29001 | PEA15 | 8682 | 200788_s_at | -0.5331 | 0.0165 | |
| GSE38129 | PEA15 | 8682 | 200788_s_at | -0.6150 | 0.0000 | |
| GSE45670 | PEA15 | 8682 | 200788_s_at | -0.4788 | 0.0039 | |
| GSE53622 | PEA15 | 8682 | 134777 | -0.5854 | 0.0000 | |
| GSE53624 | PEA15 | 8682 | 134777 | -0.4612 | 0.0000 | |
| GSE63941 | PEA15 | 8682 | 200788_s_at | -2.1234 | 0.0056 | |
| GSE77861 | PEA15 | 8682 | 200788_s_at | -0.5616 | 0.0028 | |
| GSE97050 | PEA15 | 8682 | A_24_P410952 | -0.2480 | 0.2927 | |
| SRP007169 | PEA15 | 8682 | RNAseq | -2.0931 | 0.0000 | |
| SRP008496 | PEA15 | 8682 | RNAseq | -1.6558 | 0.0000 | |
| SRP064894 | PEA15 | 8682 | RNAseq | -0.3805 | 0.0290 | |
| SRP133303 | PEA15 | 8682 | RNAseq | -0.1250 | 0.5140 | |
| SRP159526 | PEA15 | 8682 | RNAseq | -0.6527 | 0.0010 | |
| SRP193095 | PEA15 | 8682 | RNAseq | -0.6211 | 0.0009 | |
| SRP219564 | PEA15 | 8682 | RNAseq | -0.5963 | 0.0875 | |
| TCGA | PEA15 | 8682 | RNAseq | 0.0360 | 0.4643 |
Upregulated datasets: 0; Downregulated datasets: 4.
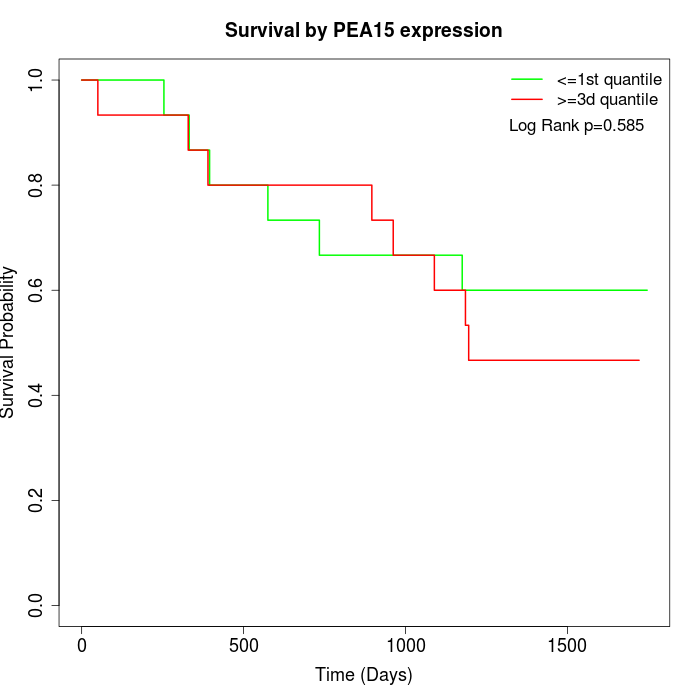
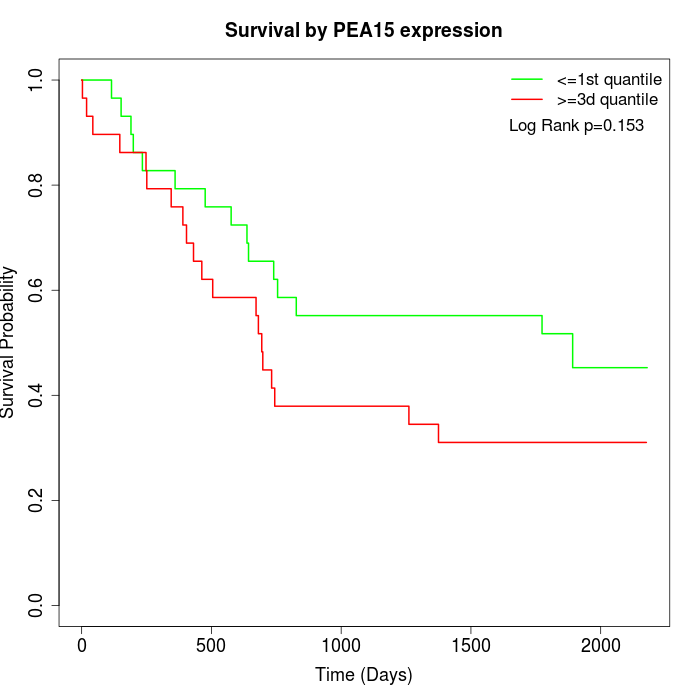
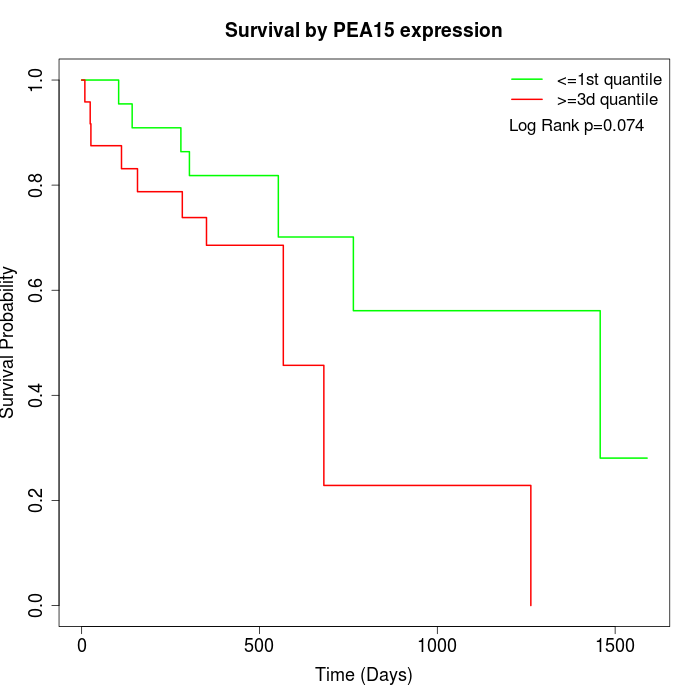
Survival by PEA15 expression:
Note: Click image to view full size file.
Copy number change of PEA15:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PEA15 | 8682 | 10 | 0 | 20 | |
| GSE20123 | PEA15 | 8682 | 10 | 0 | 20 | |
| GSE43470 | PEA15 | 8682 | 7 | 2 | 34 | |
| GSE46452 | PEA15 | 8682 | 2 | 1 | 56 | |
| GSE47630 | PEA15 | 8682 | 15 | 0 | 25 | |
| GSE54993 | PEA15 | 8682 | 0 | 5 | 65 | |
| GSE54994 | PEA15 | 8682 | 16 | 0 | 37 | |
| GSE60625 | PEA15 | 8682 | 0 | 0 | 11 | |
| GSE74703 | PEA15 | 8682 | 7 | 2 | 27 | |
| GSE74704 | PEA15 | 8682 | 4 | 0 | 16 | |
| TCGA | PEA15 | 8682 | 44 | 3 | 49 |
Total number of gains: 115; Total number of losses: 13; Total Number of normals: 360.
Somatic mutations of PEA15:
Generating mutation plots.
Highly correlated genes for PEA15:
Showing top 20/983 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PEA15 | SLC44A3 | 0.755554 | 3 | 0 | 3 |
| PEA15 | CCNYL1 | 0.732531 | 3 | 0 | 3 |
| PEA15 | SNORA68 | 0.724555 | 3 | 0 | 3 |
| PEA15 | SPNS2 | 0.716501 | 3 | 0 | 3 |
| PEA15 | CLIC3 | 0.716415 | 5 | 0 | 5 |
| PEA15 | B3GNT8 | 0.716132 | 3 | 0 | 3 |
| PEA15 | SFTA2 | 0.708223 | 3 | 0 | 3 |
| PEA15 | HOTAIRM1 | 0.7013 | 5 | 0 | 4 |
| PEA15 | STBD1 | 0.696743 | 3 | 0 | 3 |
| PEA15 | RANBP9 | 0.695603 | 7 | 0 | 6 |
| PEA15 | ZFAND2B | 0.692539 | 6 | 0 | 6 |
| PEA15 | KLHL20 | 0.690969 | 3 | 0 | 3 |
| PEA15 | PITHD1 | 0.680845 | 4 | 0 | 4 |
| PEA15 | HSPB8 | 0.68019 | 12 | 0 | 12 |
| PEA15 | GGA3 | 0.677458 | 3 | 0 | 3 |
| PEA15 | VAT1 | 0.677369 | 10 | 0 | 9 |
| PEA15 | ARHGAP10 | 0.675376 | 12 | 0 | 11 |
| PEA15 | UBE2E1 | 0.675161 | 7 | 0 | 7 |
| PEA15 | KRT78 | 0.674209 | 5 | 0 | 4 |
| PEA15 | VASN | 0.673373 | 6 | 0 | 6 |
For details and further investigation, click here