| Full name: Rho GTPase activating protein 10 | Alias Symbol: FLJ20896|FLJ41791|GRAF2 | ||
| Type: protein-coding gene | Cytoband: 4q31.23 | ||
| Entrez ID: 79658 | HGNC ID: HGNC:26099 | Ensembl Gene: ENSG00000071205 | OMIM ID: 609746 |
| Drug and gene relationship at DGIdb | |||
ARHGAP10 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa05100 | Bacterial invasion of epithelial cells |
Expression of ARHGAP10:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ARHGAP10 | 79658 | 219431_at | -1.1649 | 0.0206 | |
| GSE20347 | ARHGAP10 | 79658 | 219431_at | -1.0256 | 0.0000 | |
| GSE23400 | ARHGAP10 | 79658 | 219431_at | -0.7931 | 0.0000 | |
| GSE26886 | ARHGAP10 | 79658 | 219431_at | -1.2165 | 0.0000 | |
| GSE29001 | ARHGAP10 | 79658 | 219431_at | -1.4479 | 0.0000 | |
| GSE38129 | ARHGAP10 | 79658 | 219431_at | -1.1630 | 0.0000 | |
| GSE45670 | ARHGAP10 | 79658 | 219431_at | -0.6909 | 0.0022 | |
| GSE53622 | ARHGAP10 | 79658 | 25284 | -1.8495 | 0.0000 | |
| GSE53624 | ARHGAP10 | 79658 | 25284 | -1.7421 | 0.0000 | |
| GSE63941 | ARHGAP10 | 79658 | 219431_at | -1.0084 | 0.0011 | |
| GSE77861 | ARHGAP10 | 79658 | 219431_at | -0.7100 | 0.0014 | |
| GSE97050 | ARHGAP10 | 79658 | A_23_P21548 | -1.0539 | 0.1399 | |
| SRP007169 | ARHGAP10 | 79658 | RNAseq | -3.0003 | 0.0000 | |
| SRP008496 | ARHGAP10 | 79658 | RNAseq | -3.0177 | 0.0000 | |
| SRP064894 | ARHGAP10 | 79658 | RNAseq | -1.7976 | 0.0000 | |
| SRP133303 | ARHGAP10 | 79658 | RNAseq | -1.6615 | 0.0000 | |
| SRP159526 | ARHGAP10 | 79658 | RNAseq | -2.5589 | 0.0000 | |
| SRP193095 | ARHGAP10 | 79658 | RNAseq | -2.2335 | 0.0000 | |
| SRP219564 | ARHGAP10 | 79658 | RNAseq | -1.3517 | 0.0456 | |
| TCGA | ARHGAP10 | 79658 | RNAseq | -0.4175 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 15.
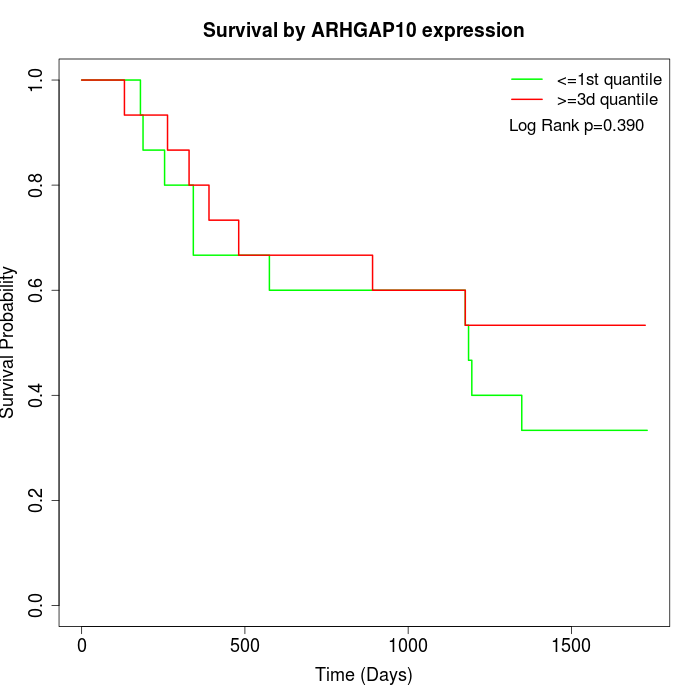
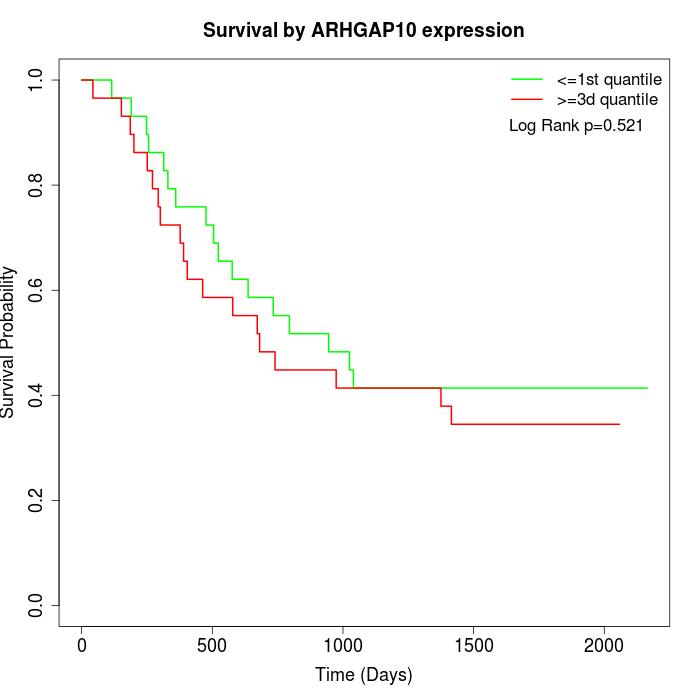
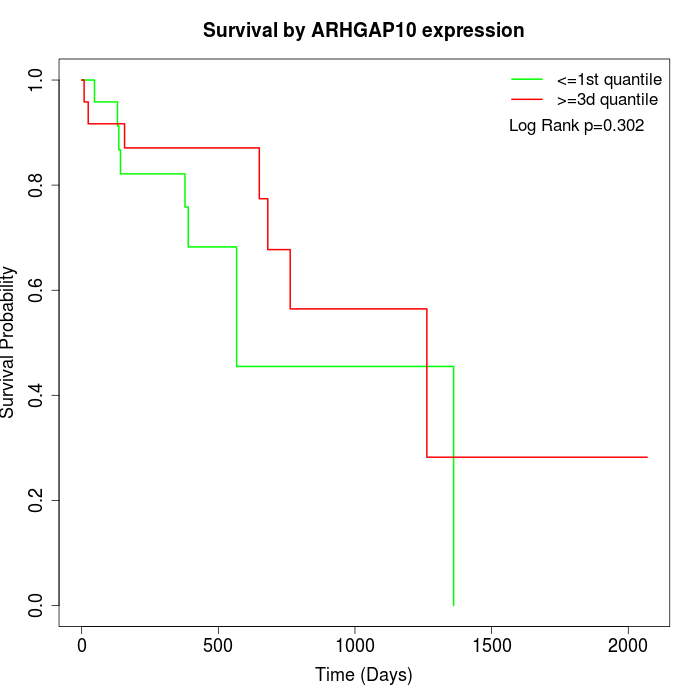
Survival by ARHGAP10 expression:
Note: Click image to view full size file.
Copy number change of ARHGAP10:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ARHGAP10 | 79658 | 0 | 12 | 18 | |
| GSE20123 | ARHGAP10 | 79658 | 0 | 12 | 18 | |
| GSE43470 | ARHGAP10 | 79658 | 0 | 13 | 30 | |
| GSE46452 | ARHGAP10 | 79658 | 1 | 36 | 22 | |
| GSE47630 | ARHGAP10 | 79658 | 0 | 22 | 18 | |
| GSE54993 | ARHGAP10 | 79658 | 10 | 0 | 60 | |
| GSE54994 | ARHGAP10 | 79658 | 2 | 11 | 40 | |
| GSE60625 | ARHGAP10 | 79658 | 0 | 1 | 10 | |
| GSE74703 | ARHGAP10 | 79658 | 0 | 11 | 25 | |
| GSE74704 | ARHGAP10 | 79658 | 0 | 7 | 13 | |
| TCGA | ARHGAP10 | 79658 | 11 | 33 | 52 |
Total number of gains: 24; Total number of losses: 158; Total Number of normals: 306.
Somatic mutations of ARHGAP10:
Generating mutation plots.
Highly correlated genes for ARHGAP10:
Showing top 20/1666 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ARHGAP10 | MYZAP | 0.872811 | 3 | 0 | 3 |
| ARHGAP10 | ABLIM3 | 0.843483 | 11 | 0 | 11 |
| ARHGAP10 | KIAA0232 | 0.818558 | 11 | 0 | 11 |
| ARHGAP10 | FCHO2 | 0.791803 | 6 | 0 | 6 |
| ARHGAP10 | CRISP3 | 0.790321 | 10 | 0 | 10 |
| ARHGAP10 | PPP1R3C | 0.789868 | 12 | 0 | 12 |
| ARHGAP10 | KAT2B | 0.788119 | 12 | 0 | 12 |
| ARHGAP10 | SNORA68 | 0.787368 | 4 | 0 | 4 |
| ARHGAP10 | MGLL | 0.786732 | 12 | 0 | 12 |
| ARHGAP10 | CITED2 | 0.784264 | 12 | 0 | 12 |
| ARHGAP10 | PAQR8 | 0.783451 | 6 | 0 | 6 |
| ARHGAP10 | MMEL1 | 0.783304 | 3 | 0 | 3 |
| ARHGAP10 | HSPB8 | 0.783137 | 12 | 0 | 12 |
| ARHGAP10 | GYS2 | 0.782282 | 10 | 0 | 9 |
| ARHGAP10 | SH3BGRL2 | 0.781527 | 7 | 0 | 7 |
| ARHGAP10 | SASH1 | 0.780756 | 11 | 0 | 11 |
| ARHGAP10 | EIF4E3 | 0.778464 | 7 | 0 | 7 |
| ARHGAP10 | RBFOX3 | 0.778305 | 3 | 0 | 3 |
| ARHGAP10 | FAM214A | 0.777023 | 6 | 0 | 6 |
| ARHGAP10 | SFTA2 | 0.776699 | 4 | 0 | 3 |
For details and further investigation, click here