| Full name: platelet and endothelial cell adhesion molecule 1 | Alias Symbol: CD31 | ||
| Type: protein-coding gene | Cytoband: 17q23.3 | ||
| Entrez ID: 5175 | HGNC ID: HGNC:8823 | Ensembl Gene: ENSG00000261371 | OMIM ID: 173445 |
| Drug and gene relationship at DGIdb | |||
PECAM1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04670 | Leukocyte transendothelial migration |
Expression of PECAM1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PECAM1 | 5175 | 208982_at | -0.5148 | 0.5231 | |
| GSE20347 | PECAM1 | 5175 | 208982_at | 0.0498 | 0.8588 | |
| GSE23400 | PECAM1 | 5175 | 208982_at | -0.0220 | 0.8760 | |
| GSE26886 | PECAM1 | 5175 | 208982_at | 0.3199 | 0.3985 | |
| GSE29001 | PECAM1 | 5175 | 208982_at | -0.0513 | 0.9165 | |
| GSE38129 | PECAM1 | 5175 | 208982_at | -0.3719 | 0.3724 | |
| GSE45670 | PECAM1 | 5175 | 208982_at | -0.8948 | 0.0202 | |
| GSE53622 | PECAM1 | 5175 | 66484 | -0.5568 | 0.0000 | |
| GSE53624 | PECAM1 | 5175 | 14483 | -0.6327 | 0.0000 | |
| GSE63941 | PECAM1 | 5175 | 1559921_at | 0.0902 | 0.7018 | |
| GSE77861 | PECAM1 | 5175 | 208982_at | 0.6844 | 0.1236 | |
| GSE97050 | PECAM1 | 5175 | A_33_P3229402 | 0.5608 | 0.1889 | |
| SRP007169 | PECAM1 | 5175 | RNAseq | 1.9664 | 0.0001 | |
| SRP008496 | PECAM1 | 5175 | RNAseq | 2.1209 | 0.0000 | |
| SRP064894 | PECAM1 | 5175 | RNAseq | 0.9542 | 0.0067 | |
| SRP133303 | PECAM1 | 5175 | RNAseq | -0.3019 | 0.2000 | |
| SRP159526 | PECAM1 | 5175 | RNAseq | 0.3758 | 0.4829 | |
| SRP193095 | PECAM1 | 5175 | RNAseq | 0.9757 | 0.0002 | |
| SRP219564 | PECAM1 | 5175 | RNAseq | 0.1602 | 0.7770 | |
| TCGA | PECAM1 | 5175 | RNAseq | -0.3541 | 0.0001 |
Upregulated datasets: 2; Downregulated datasets: 0.
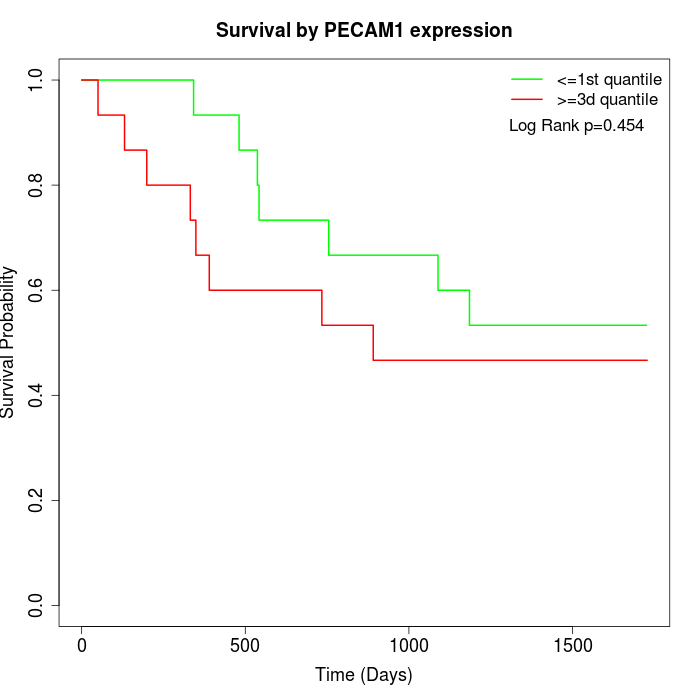
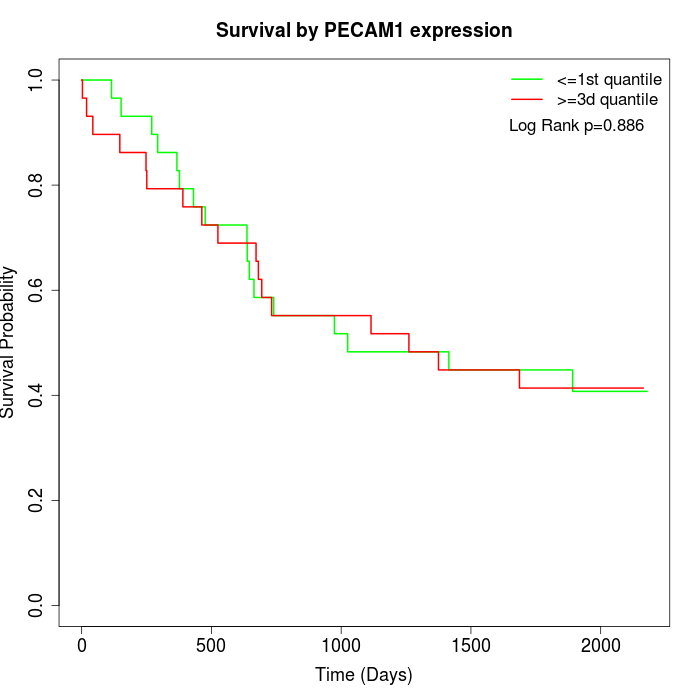
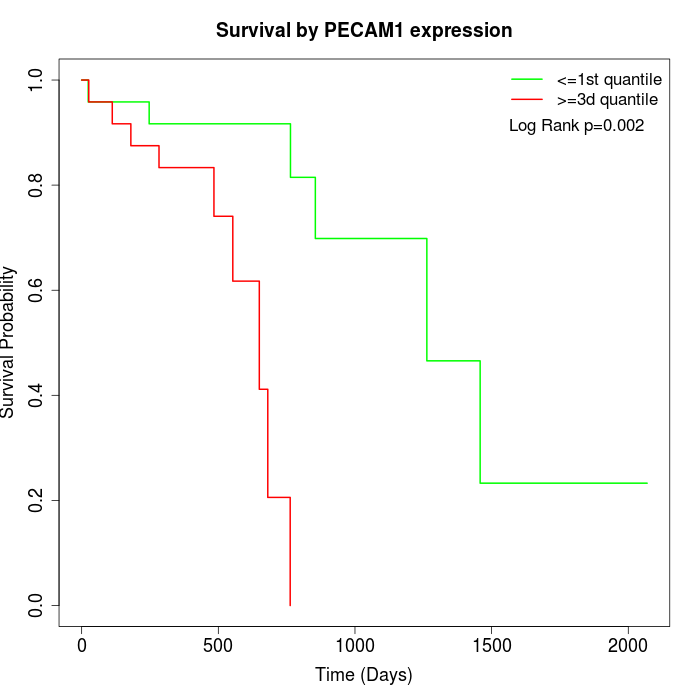
Survival by PECAM1 expression:
Note: Click image to view full size file.
Copy number change of PECAM1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PECAM1 | 5175 | 3 | 1 | 26 | |
| GSE20123 | PECAM1 | 5175 | 3 | 1 | 26 | |
| GSE43470 | PECAM1 | 5175 | 4 | 0 | 39 | |
| GSE46452 | PECAM1 | 5175 | 32 | 0 | 27 | |
| GSE47630 | PECAM1 | 5175 | 7 | 1 | 32 | |
| GSE54993 | PECAM1 | 5175 | 2 | 5 | 63 | |
| GSE54994 | PECAM1 | 5175 | 9 | 4 | 40 | |
| GSE60625 | PECAM1 | 5175 | 4 | 0 | 7 | |
| GSE74703 | PECAM1 | 5175 | 4 | 0 | 32 | |
| GSE74704 | PECAM1 | 5175 | 3 | 1 | 16 |
Total number of gains: 71; Total number of losses: 13; Total Number of normals: 308.
Somatic mutations of PECAM1:
Generating mutation plots.
Highly correlated genes for PECAM1:
Showing top 20/552 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PECAM1 | GNG11 | 0.747163 | 12 | 0 | 11 |
| PECAM1 | CD93 | 0.743969 | 12 | 0 | 10 |
| PECAM1 | COL15A1 | 0.737059 | 11 | 0 | 10 |
| PECAM1 | CLEC14A | 0.730205 | 7 | 0 | 7 |
| PECAM1 | A2M | 0.730099 | 12 | 0 | 10 |
| PECAM1 | C3 | 0.726227 | 10 | 0 | 10 |
| PECAM1 | NCOR2 | 0.707633 | 3 | 0 | 3 |
| PECAM1 | MGRN1 | 0.707435 | 3 | 0 | 3 |
| PECAM1 | VEGFB | 0.705769 | 3 | 0 | 3 |
| PECAM1 | ECSCR | 0.70218 | 9 | 0 | 8 |
| PECAM1 | SSH1 | 0.701817 | 3 | 0 | 3 |
| PECAM1 | TFPI | 0.699122 | 10 | 0 | 9 |
| PECAM1 | TEK | 0.697538 | 7 | 0 | 5 |
| PECAM1 | CDH5 | 0.695716 | 13 | 0 | 10 |
| PECAM1 | TIE1 | 0.695054 | 10 | 0 | 9 |
| PECAM1 | XPNPEP2 | 0.694506 | 5 | 0 | 5 |
| PECAM1 | SYN1 | 0.691051 | 3 | 0 | 3 |
| PECAM1 | ARHGAP15 | 0.68834 | 4 | 0 | 4 |
| PECAM1 | ZNF566 | 0.683959 | 3 | 0 | 3 |
| PECAM1 | EDNRB | 0.683408 | 8 | 0 | 7 |
For details and further investigation, click here