| Full name: complement C3 | Alias Symbol: CPAMD1|ARMD9|C3a|C3b | ||
| Type: protein-coding gene | Cytoband: 19p13.3 | ||
| Entrez ID: 718 | HGNC ID: HGNC:1318 | Ensembl Gene: ENSG00000125730 | OMIM ID: 120700 |
| Drug and gene relationship at DGIdb | |||
C3 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04610 | Complement and coagulation cascades | |
| hsa05133 | Pertussis | |
| hsa05140 | Leishmaniasis | |
| hsa05150 | Staphylococcus aureus infection | |
| hsa05152 | Tuberculosis |
Expression of C3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | C3 | 718 | 217767_at | -0.5120 | 0.6600 | |
| GSE20347 | C3 | 718 | 217767_at | 0.3194 | 0.4166 | |
| GSE23400 | C3 | 718 | 217767_at | -0.3530 | 0.1203 | |
| GSE26886 | C3 | 718 | 217767_at | 1.8773 | 0.0191 | |
| GSE29001 | C3 | 718 | 217767_at | 0.5541 | 0.3888 | |
| GSE38129 | C3 | 718 | 217767_at | -0.2676 | 0.6236 | |
| GSE45670 | C3 | 718 | 217767_at | -2.6129 | 0.0000 | |
| GSE53622 | C3 | 718 | 5914 | -0.7473 | 0.0000 | |
| GSE53624 | C3 | 718 | 76778 | -0.6495 | 0.0000 | |
| GSE63941 | C3 | 718 | 217767_at | -0.4298 | 0.8073 | |
| GSE77861 | C3 | 718 | 217767_at | 0.8114 | 0.0242 | |
| GSE97050 | C3 | 718 | A_33_P3347869 | 0.3493 | 0.6048 | |
| SRP007169 | C3 | 718 | RNAseq | 3.9220 | 0.0000 | |
| SRP008496 | C3 | 718 | RNAseq | 4.0967 | 0.0000 | |
| SRP064894 | C3 | 718 | RNAseq | 1.1415 | 0.0391 | |
| SRP133303 | C3 | 718 | RNAseq | -0.4290 | 0.2220 | |
| SRP159526 | C3 | 718 | RNAseq | -0.4087 | 0.4605 | |
| SRP193095 | C3 | 718 | RNAseq | 1.6437 | 0.0006 | |
| SRP219564 | C3 | 718 | RNAseq | 1.0941 | 0.2588 | |
| TCGA | C3 | 718 | RNAseq | -0.4073 | 0.0031 |
Upregulated datasets: 5; Downregulated datasets: 1.
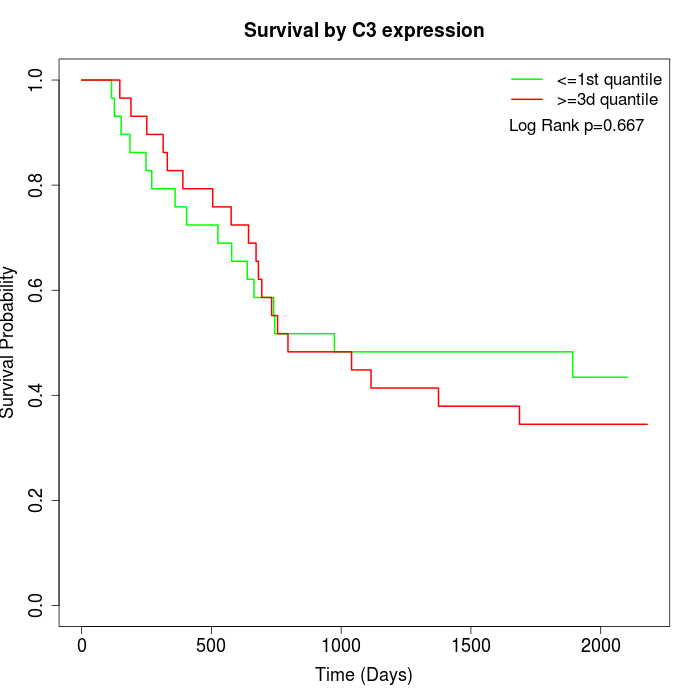
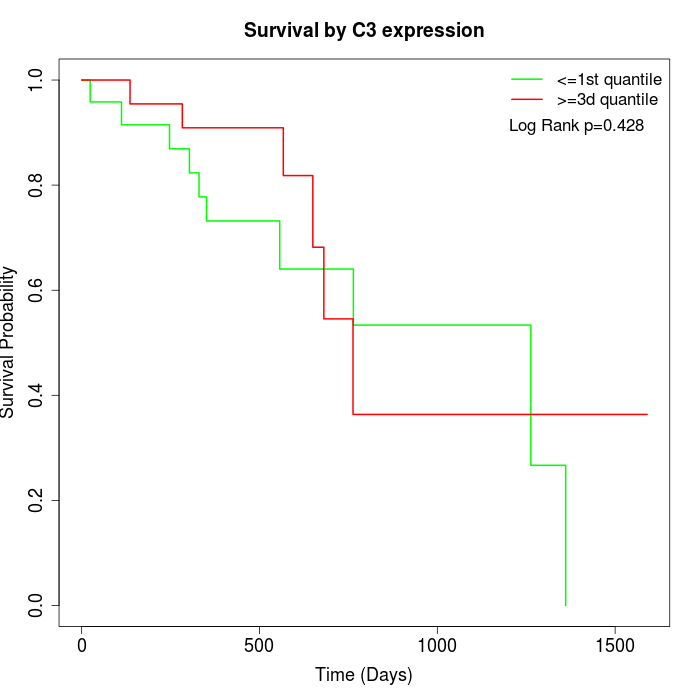
Survival by C3 expression:
Note: Click image to view full size file.
Copy number change of C3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | C3 | 718 | 5 | 3 | 22 | |
| GSE20123 | C3 | 718 | 4 | 2 | 24 | |
| GSE43470 | C3 | 718 | 2 | 6 | 35 | |
| GSE46452 | C3 | 718 | 47 | 1 | 11 | |
| GSE47630 | C3 | 718 | 5 | 7 | 28 | |
| GSE54993 | C3 | 718 | 16 | 3 | 51 | |
| GSE54994 | C3 | 718 | 6 | 14 | 33 | |
| GSE60625 | C3 | 718 | 9 | 0 | 2 | |
| GSE74703 | C3 | 718 | 2 | 4 | 30 | |
| GSE74704 | C3 | 718 | 1 | 2 | 17 | |
| TCGA | C3 | 718 | 9 | 19 | 68 |
Total number of gains: 106; Total number of losses: 61; Total Number of normals: 321.
Somatic mutations of C3:
Generating mutation plots.
Highly correlated genes for C3:
Showing top 20/749 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| C3 | LINC01279 | 0.775824 | 3 | 0 | 3 |
| C3 | F13A1 | 0.735789 | 8 | 0 | 7 |
| C3 | PECAM1 | 0.726227 | 10 | 0 | 10 |
| C3 | GNG2 | 0.716916 | 4 | 0 | 4 |
| C3 | MEF2C | 0.710569 | 9 | 0 | 8 |
| C3 | GIMAP1 | 0.708856 | 5 | 0 | 5 |
| C3 | FBLN5 | 0.702423 | 9 | 0 | 8 |
| C3 | CD34 | 0.701667 | 6 | 0 | 6 |
| C3 | STAB1 | 0.700125 | 10 | 0 | 9 |
| C3 | GIMAP5 | 0.699414 | 3 | 0 | 3 |
| C3 | DAB2 | 0.697552 | 9 | 0 | 9 |
| C3 | RNASE1 | 0.69463 | 10 | 0 | 10 |
| C3 | PMP22 | 0.691526 | 9 | 0 | 8 |
| C3 | ENPP2 | 0.690716 | 7 | 0 | 7 |
| C3 | C1orf54 | 0.686571 | 8 | 0 | 7 |
| C3 | ZEB2 | 0.685988 | 9 | 0 | 9 |
| C3 | FGF7 | 0.685527 | 7 | 0 | 7 |
| C3 | IGF1 | 0.684665 | 9 | 0 | 7 |
| C3 | CCDC88A | 0.681631 | 3 | 0 | 3 |
| C3 | SLC46A3 | 0.681338 | 7 | 0 | 7 |
For details and further investigation, click here