| Full name: period circadian regulator 2 | Alias Symbol: KIAA0347 | ||
| Type: protein-coding gene | Cytoband: 2q37.3 | ||
| Entrez ID: 8864 | HGNC ID: HGNC:8846 | Ensembl Gene: ENSG00000132326 | OMIM ID: 603426 |
| Drug and gene relationship at DGIdb | |||
PER2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04710 | Circadian rhythm | |
| hsa04713 | Circadian entrainment |
Expression of PER2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PER2 | 8864 | 205251_at | -0.3482 | 0.8263 | |
| GSE20347 | PER2 | 8864 | 205251_at | -1.2079 | 0.0002 | |
| GSE23400 | PER2 | 8864 | 205251_at | -1.0722 | 0.0000 | |
| GSE26886 | PER2 | 8864 | 205251_at | -0.2985 | 0.2577 | |
| GSE29001 | PER2 | 8864 | 205251_at | -1.1842 | 0.0004 | |
| GSE38129 | PER2 | 8864 | 205251_at | -1.0465 | 0.0000 | |
| GSE45670 | PER2 | 8864 | 205251_at | -0.4874 | 0.1436 | |
| GSE53622 | PER2 | 8864 | 64823 | -1.0566 | 0.0000 | |
| GSE53624 | PER2 | 8864 | 64296 | -1.6408 | 0.0000 | |
| GSE63941 | PER2 | 8864 | 205251_at | 1.5518 | 0.0366 | |
| GSE77861 | PER2 | 8864 | 205251_at | 0.0669 | 0.8768 | |
| GSE97050 | PER2 | 8864 | A_23_P411162 | -0.2885 | 0.5619 | |
| SRP007169 | PER2 | 8864 | RNAseq | -0.2456 | 0.5510 | |
| SRP008496 | PER2 | 8864 | RNAseq | -0.3051 | 0.3749 | |
| SRP064894 | PER2 | 8864 | RNAseq | -1.7360 | 0.0000 | |
| SRP133303 | PER2 | 8864 | RNAseq | -0.6217 | 0.0036 | |
| SRP159526 | PER2 | 8864 | RNAseq | -0.6339 | 0.2037 | |
| SRP193095 | PER2 | 8864 | RNAseq | -0.3971 | 0.1096 | |
| SRP219564 | PER2 | 8864 | RNAseq | -0.0924 | 0.8787 | |
| TCGA | PER2 | 8864 | RNAseq | -0.3473 | 0.0000 |
Upregulated datasets: 1; Downregulated datasets: 7.
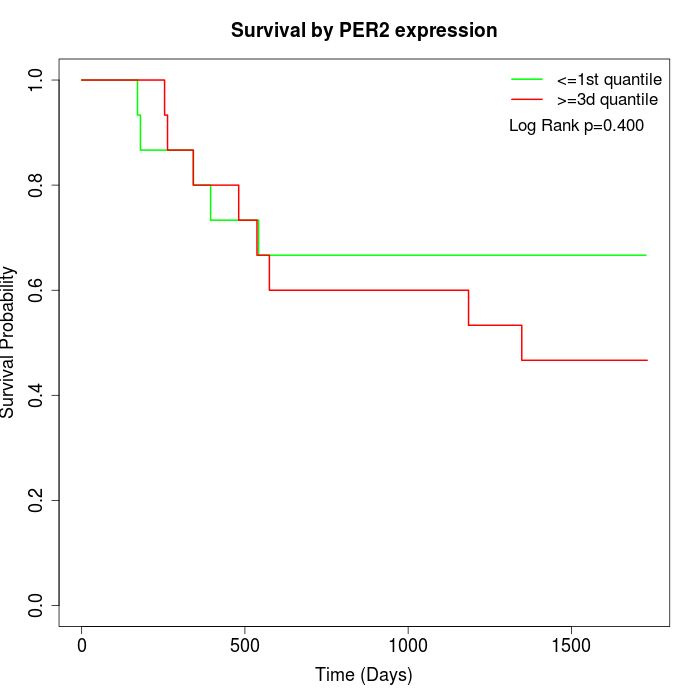
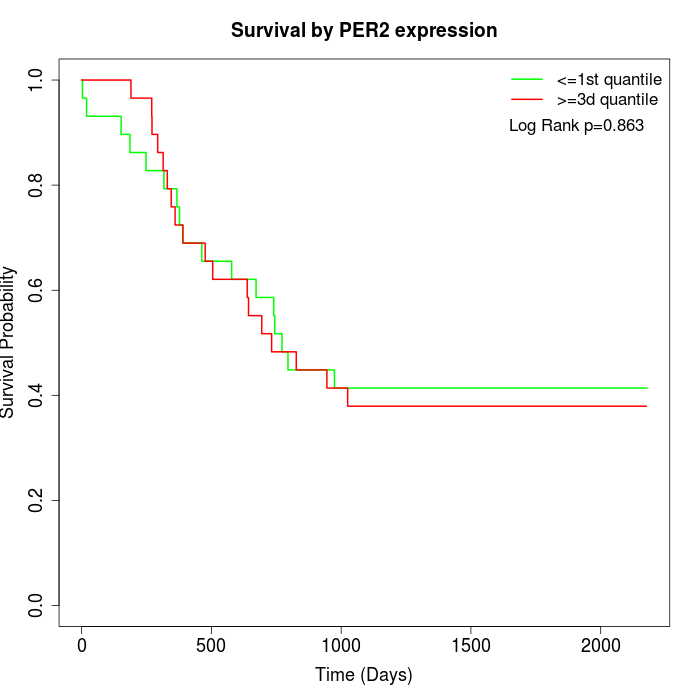
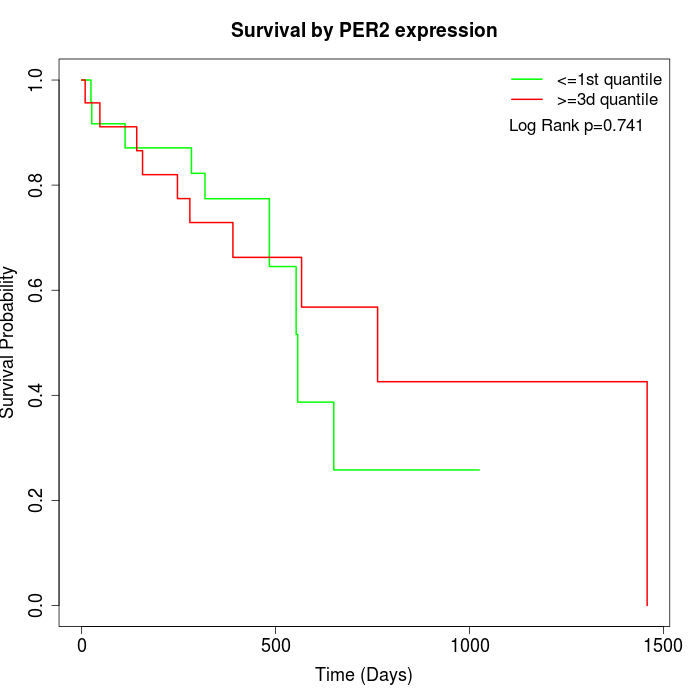
Survival by PER2 expression:
Note: Click image to view full size file.
Copy number change of PER2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PER2 | 8864 | 1 | 12 | 17 | |
| GSE20123 | PER2 | 8864 | 1 | 12 | 17 | |
| GSE43470 | PER2 | 8864 | 1 | 9 | 33 | |
| GSE46452 | PER2 | 8864 | 0 | 5 | 54 | |
| GSE47630 | PER2 | 8864 | 4 | 5 | 31 | |
| GSE54993 | PER2 | 8864 | 3 | 2 | 65 | |
| GSE54994 | PER2 | 8864 | 8 | 9 | 36 | |
| GSE60625 | PER2 | 8864 | 0 | 3 | 8 | |
| GSE74703 | PER2 | 8864 | 1 | 7 | 28 | |
| GSE74704 | PER2 | 8864 | 1 | 5 | 14 | |
| TCGA | PER2 | 8864 | 12 | 27 | 57 |
Total number of gains: 32; Total number of losses: 96; Total Number of normals: 360.
Somatic mutations of PER2:
Generating mutation plots.
Highly correlated genes for PER2:
Showing top 20/467 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PER2 | LYSMD1 | 0.727449 | 3 | 0 | 3 |
| PER2 | KCNB1 | 0.68815 | 3 | 0 | 3 |
| PER2 | TRAF3IP1 | 0.680569 | 4 | 0 | 4 |
| PER2 | C1QTNF7 | 0.669957 | 3 | 0 | 3 |
| PER2 | EIF4E3 | 0.66415 | 3 | 0 | 3 |
| PER2 | CLEC3B | 0.662459 | 3 | 0 | 3 |
| PER2 | XKR4 | 0.658913 | 3 | 0 | 3 |
| PER2 | ETFDH | 0.656246 | 6 | 0 | 6 |
| PER2 | HPSE2 | 0.654325 | 3 | 0 | 3 |
| PER2 | TCP11L2 | 0.648521 | 3 | 0 | 3 |
| PER2 | RMND5B | 0.648406 | 7 | 0 | 7 |
| PER2 | UBL3 | 0.645386 | 7 | 0 | 6 |
| PER2 | PITHD1 | 0.64411 | 3 | 0 | 3 |
| PER2 | PAAF1 | 0.641959 | 4 | 0 | 3 |
| PER2 | RANBP9 | 0.639468 | 6 | 0 | 6 |
| PER2 | RAB11A | 0.639417 | 8 | 0 | 7 |
| PER2 | CGNL1 | 0.637159 | 3 | 0 | 3 |
| PER2 | KANK1 | 0.636879 | 7 | 0 | 7 |
| PER2 | FHL5 | 0.635193 | 4 | 0 | 3 |
| PER2 | FCER1A | 0.634823 | 7 | 0 | 7 |
For details and further investigation, click here