| Full name: post-GPI attachment to proteins 2 | Alias Symbol: FRAG1|CWH43-N | ||
| Type: protein-coding gene | Cytoband: 11p15.4 | ||
| Entrez ID: 27315 | HGNC ID: HGNC:17893 | Ensembl Gene: ENSG00000148985 | OMIM ID: 615187 |
| Drug and gene relationship at DGIdb | |||
Expression of PGAP2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PGAP2 | 27315 | 215293_s_at | -0.1729 | 0.6880 | |
| GSE20347 | PGAP2 | 27315 | 215293_s_at | -0.3103 | 0.0620 | |
| GSE23400 | PGAP2 | 27315 | 215293_s_at | -0.0521 | 0.3596 | |
| GSE26886 | PGAP2 | 27315 | 215293_s_at | -0.6448 | 0.0007 | |
| GSE29001 | PGAP2 | 27315 | 41858_at | -0.3538 | 0.3117 | |
| GSE38129 | PGAP2 | 27315 | 41858_at | 0.0168 | 0.9274 | |
| GSE45670 | PGAP2 | 27315 | 41858_at | 0.1053 | 0.5064 | |
| GSE53622 | PGAP2 | 27315 | 70211 | -0.0494 | 0.5193 | |
| GSE53624 | PGAP2 | 27315 | 70211 | -0.2992 | 0.0000 | |
| GSE63941 | PGAP2 | 27315 | 41858_at | 0.5060 | 0.1309 | |
| GSE77861 | PGAP2 | 27315 | 41858_at | -0.1552 | 0.6551 | |
| GSE97050 | PGAP2 | 27315 | A_23_P64560 | 0.1279 | 0.5619 | |
| SRP007169 | PGAP2 | 27315 | RNAseq | -0.3708 | 0.4184 | |
| SRP008496 | PGAP2 | 27315 | RNAseq | -0.1638 | 0.6068 | |
| SRP064894 | PGAP2 | 27315 | RNAseq | -0.3694 | 0.0652 | |
| SRP133303 | PGAP2 | 27315 | RNAseq | -0.7973 | 0.0004 | |
| SRP159526 | PGAP2 | 27315 | RNAseq | -0.6137 | 0.0440 | |
| SRP193095 | PGAP2 | 27315 | RNAseq | -0.5578 | 0.0003 | |
| SRP219564 | PGAP2 | 27315 | RNAseq | -0.3978 | 0.4987 | |
| TCGA | PGAP2 | 27315 | RNAseq | 0.1196 | 0.0936 |
Upregulated datasets: 0; Downregulated datasets: 0.
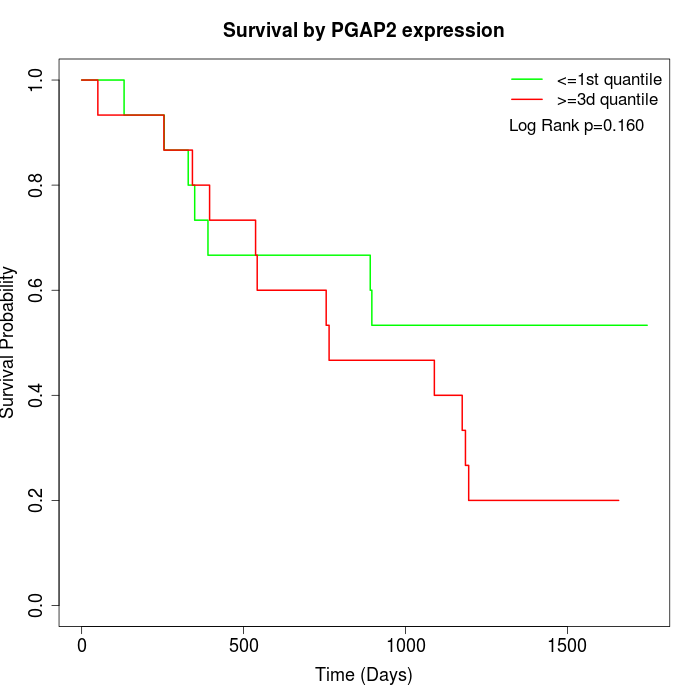
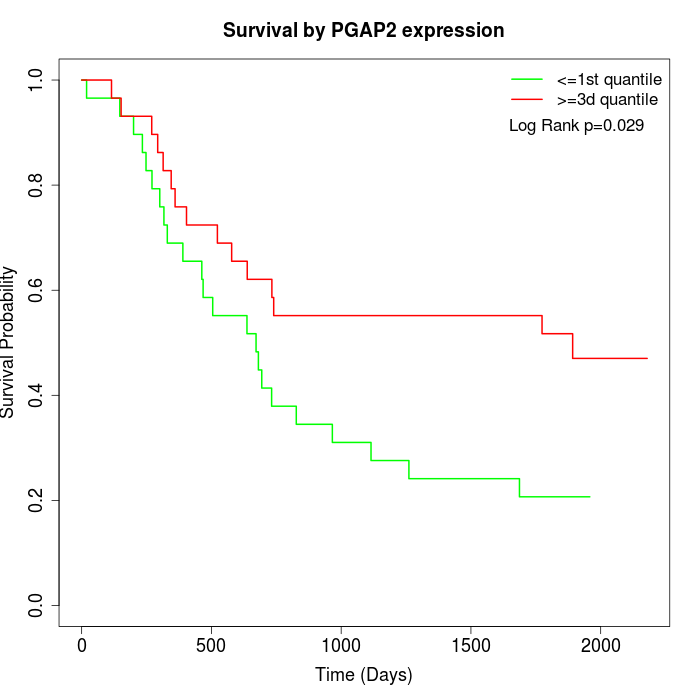
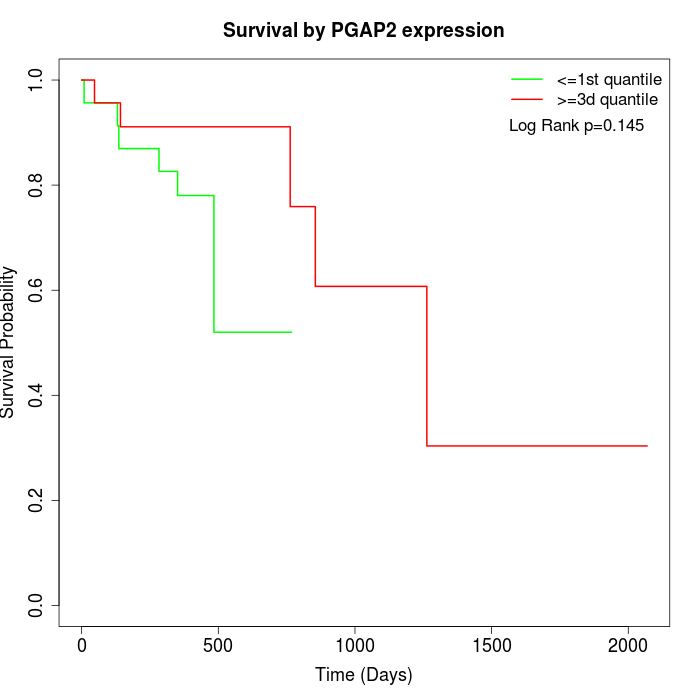
Survival by PGAP2 expression:
Note: Click image to view full size file.
Copy number change of PGAP2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PGAP2 | 27315 | 1 | 12 | 17 | |
| GSE20123 | PGAP2 | 27315 | 1 | 11 | 18 | |
| GSE43470 | PGAP2 | 27315 | 1 | 9 | 33 | |
| GSE46452 | PGAP2 | 27315 | 7 | 8 | 44 | |
| GSE47630 | PGAP2 | 27315 | 4 | 12 | 24 | |
| GSE54993 | PGAP2 | 27315 | 3 | 1 | 66 | |
| GSE54994 | PGAP2 | 27315 | 1 | 12 | 40 | |
| GSE60625 | PGAP2 | 27315 | 0 | 0 | 11 | |
| GSE74703 | PGAP2 | 27315 | 1 | 8 | 27 | |
| GSE74704 | PGAP2 | 27315 | 0 | 8 | 12 | |
| TCGA | PGAP2 | 27315 | 7 | 38 | 51 |
Total number of gains: 26; Total number of losses: 119; Total Number of normals: 343.
Somatic mutations of PGAP2:
Generating mutation plots.
Highly correlated genes for PGAP2:
Showing top 20/363 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PGAP2 | SLC35A4 | 0.728691 | 3 | 0 | 3 |
| PGAP2 | NCOR1 | 0.727557 | 3 | 0 | 3 |
| PGAP2 | ABHD14B | 0.727197 | 3 | 0 | 3 |
| PGAP2 | TADA3 | 0.726499 | 3 | 0 | 3 |
| PGAP2 | ACER3 | 0.709695 | 3 | 0 | 3 |
| PGAP2 | TRERF1 | 0.706923 | 3 | 0 | 3 |
| PGAP2 | SYAP1 | 0.701931 | 3 | 0 | 3 |
| PGAP2 | PKP2 | 0.697271 | 3 | 0 | 3 |
| PGAP2 | PDLIM1 | 0.697087 | 4 | 0 | 3 |
| PGAP2 | VPS25 | 0.68728 | 3 | 0 | 3 |
| PGAP2 | SRP68 | 0.681108 | 3 | 0 | 3 |
| PGAP2 | MCU | 0.678035 | 3 | 0 | 3 |
| PGAP2 | ATP11B | 0.676376 | 4 | 0 | 3 |
| PGAP2 | TBC1D2 | 0.670141 | 4 | 0 | 3 |
| PGAP2 | NDUFAF3 | 0.669779 | 3 | 0 | 3 |
| PGAP2 | SNX17 | 0.666725 | 3 | 0 | 3 |
| PGAP2 | SCAMP2 | 0.66034 | 5 | 0 | 3 |
| PGAP2 | OR7E24 | 0.658919 | 3 | 0 | 3 |
| PGAP2 | MUL1 | 0.655453 | 3 | 0 | 3 |
| PGAP2 | POLR2L | 0.653983 | 5 | 0 | 4 |
For details and further investigation, click here