| Full name: chitinase 1 | Alias Symbol: CHIT|CHI3 | ||
| Type: protein-coding gene | Cytoband: 1q32.1 | ||
| Entrez ID: 1118 | HGNC ID: HGNC:1936 | Ensembl Gene: ENSG00000133063 | OMIM ID: 600031 |
| Drug and gene relationship at DGIdb | |||
Expression of CHIT1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CHIT1 | 1118 | 208168_s_at | 0.5934 | 0.5458 | |
| GSE20347 | CHIT1 | 1118 | 208168_s_at | 0.1475 | 0.1683 | |
| GSE23400 | CHIT1 | 1118 | 208168_s_at | 0.0624 | 0.3861 | |
| GSE26886 | CHIT1 | 1118 | 208168_s_at | 0.2495 | 0.1490 | |
| GSE29001 | CHIT1 | 1118 | 208168_s_at | -0.1319 | 0.3355 | |
| GSE38129 | CHIT1 | 1118 | 208168_s_at | 0.2417 | 0.0790 | |
| GSE45670 | CHIT1 | 1118 | 208168_s_at | 0.1551 | 0.1003 | |
| GSE53622 | CHIT1 | 1118 | 72994 | 1.0389 | 0.0000 | |
| GSE53624 | CHIT1 | 1118 | 72994 | 0.5251 | 0.0000 | |
| GSE63941 | CHIT1 | 1118 | 208168_s_at | -0.0821 | 0.5631 | |
| GSE77861 | CHIT1 | 1118 | 208168_s_at | 0.0190 | 0.8666 | |
| GSE97050 | CHIT1 | 1118 | A_33_P3388501 | 0.1673 | 0.4265 | |
| SRP133303 | CHIT1 | 1118 | RNAseq | 2.6547 | 0.0000 | |
| TCGA | CHIT1 | 1118 | RNAseq | 2.2865 | 0.0000 |
Upregulated datasets: 3; Downregulated datasets: 0.
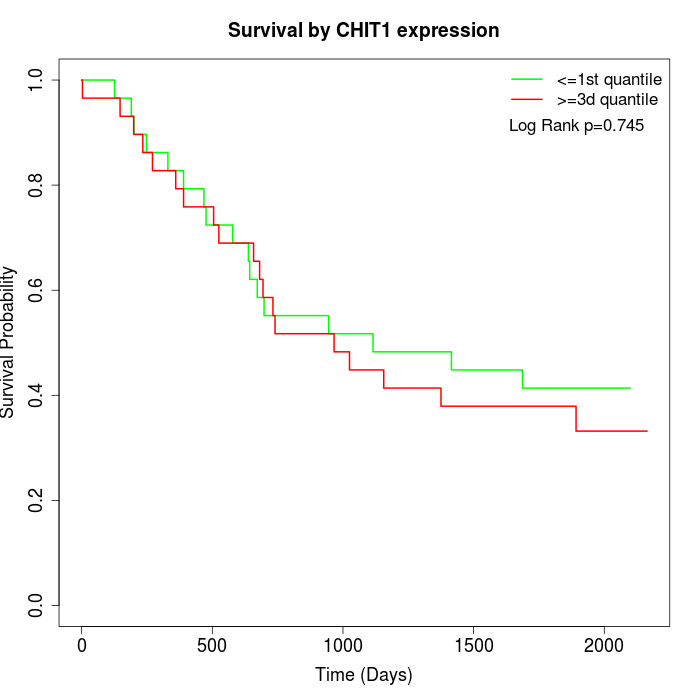
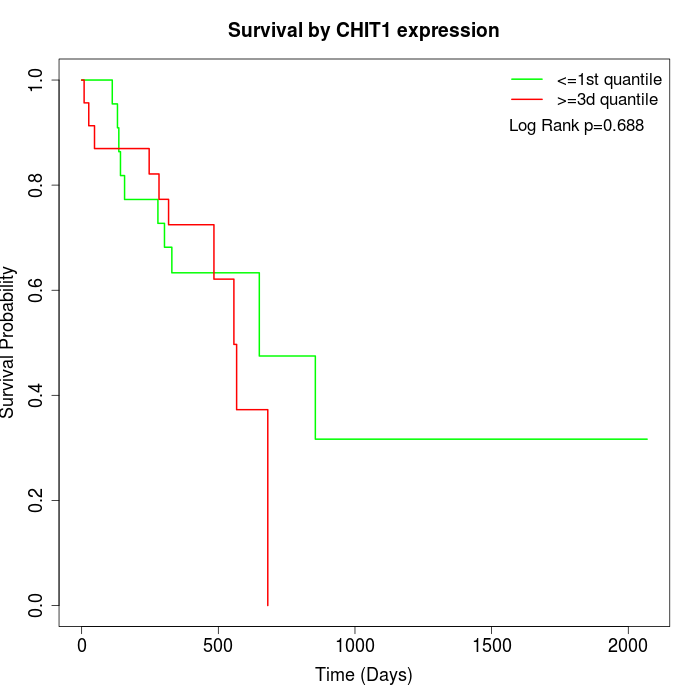
Survival by CHIT1 expression:
Note: Click image to view full size file.
Copy number change of CHIT1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CHIT1 | 1118 | 11 | 0 | 19 | |
| GSE20123 | CHIT1 | 1118 | 11 | 0 | 19 | |
| GSE43470 | CHIT1 | 1118 | 6 | 0 | 37 | |
| GSE46452 | CHIT1 | 1118 | 3 | 1 | 55 | |
| GSE47630 | CHIT1 | 1118 | 14 | 0 | 26 | |
| GSE54993 | CHIT1 | 1118 | 0 | 6 | 64 | |
| GSE54994 | CHIT1 | 1118 | 15 | 0 | 38 | |
| GSE60625 | CHIT1 | 1118 | 0 | 0 | 11 | |
| GSE74703 | CHIT1 | 1118 | 6 | 0 | 30 | |
| GSE74704 | CHIT1 | 1118 | 5 | 0 | 15 | |
| TCGA | CHIT1 | 1118 | 44 | 4 | 48 |
Total number of gains: 115; Total number of losses: 11; Total Number of normals: 362.
Somatic mutations of CHIT1:
Generating mutation plots.
Highly correlated genes for CHIT1:
Showing top 20/188 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CHIT1 | PGC | 0.801402 | 3 | 0 | 3 |
| CHIT1 | ZNF335 | 0.794657 | 3 | 0 | 3 |
| CHIT1 | NKPD1 | 0.773474 | 3 | 0 | 3 |
| CHIT1 | SMCP | 0.756011 | 3 | 0 | 3 |
| CHIT1 | KLHL35 | 0.728874 | 3 | 0 | 3 |
| CHIT1 | SLC4A1 | 0.708184 | 3 | 0 | 3 |
| CHIT1 | KCNAB3 | 0.705524 | 3 | 0 | 3 |
| CHIT1 | GPR173 | 0.700972 | 3 | 0 | 3 |
| CHIT1 | BATF2 | 0.69634 | 4 | 0 | 4 |
| CHIT1 | REG3A | 0.687962 | 3 | 0 | 3 |
| CHIT1 | GFRA4 | 0.685612 | 5 | 0 | 5 |
| CHIT1 | ACSBG2 | 0.682341 | 3 | 0 | 3 |
| CHIT1 | MYOZ3 | 0.666709 | 3 | 0 | 3 |
| CHIT1 | HGD | 0.66633 | 4 | 0 | 3 |
| CHIT1 | HRH2 | 0.662599 | 3 | 0 | 3 |
| CHIT1 | GABRG2 | 0.661504 | 3 | 0 | 3 |
| CHIT1 | GREB1 | 0.661265 | 4 | 0 | 3 |
| CHIT1 | ZFAT | 0.658316 | 3 | 0 | 3 |
| CHIT1 | HECW1 | 0.654326 | 3 | 0 | 3 |
| CHIT1 | LRFN1 | 0.654127 | 4 | 0 | 3 |
For details and further investigation, click here