| Full name: protein inhibitor of activated STAT 4 | Alias Symbol: Piasg|PIASY|FLJ12419|ZMIZ6 | ||
| Type: protein-coding gene | Cytoband: 19p13.3 | ||
| Entrez ID: 51588 | HGNC ID: HGNC:17002 | Ensembl Gene: ENSG00000105229 | OMIM ID: 605989 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
PIAS4 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04630 | Jak-STAT signaling pathway |
Expression of PIAS4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PIAS4 | 51588 | 212879_x_at | 0.3225 | 0.2369 | |
| GSE20347 | PIAS4 | 51588 | 212879_x_at | 0.1079 | 0.1309 | |
| GSE23400 | PIAS4 | 51588 | 212879_x_at | -0.0285 | 0.4666 | |
| GSE26886 | PIAS4 | 51588 | 212881_at | 1.3017 | 0.0000 | |
| GSE29001 | PIAS4 | 51588 | 212881_at | 0.6565 | 0.0056 | |
| GSE38129 | PIAS4 | 51588 | 212879_x_at | 0.2121 | 0.0026 | |
| GSE45670 | PIAS4 | 51588 | 212881_at | 0.5862 | 0.0001 | |
| GSE53622 | PIAS4 | 51588 | 35575 | 0.2532 | 0.0005 | |
| GSE53624 | PIAS4 | 51588 | 35575 | 0.3283 | 0.0000 | |
| GSE63941 | PIAS4 | 51588 | 212881_at | 0.2697 | 0.4531 | |
| GSE77861 | PIAS4 | 51588 | 212879_x_at | 0.2570 | 0.0587 | |
| GSE97050 | PIAS4 | 51588 | A_33_P3260062 | 0.1209 | 0.6603 | |
| SRP007169 | PIAS4 | 51588 | RNAseq | -0.2146 | 0.5882 | |
| SRP008496 | PIAS4 | 51588 | RNAseq | -0.5400 | 0.0852 | |
| SRP064894 | PIAS4 | 51588 | RNAseq | 0.4964 | 0.0130 | |
| SRP133303 | PIAS4 | 51588 | RNAseq | 0.0732 | 0.6460 | |
| SRP159526 | PIAS4 | 51588 | RNAseq | 0.2063 | 0.3535 | |
| SRP193095 | PIAS4 | 51588 | RNAseq | 0.3179 | 0.0010 | |
| SRP219564 | PIAS4 | 51588 | RNAseq | 0.1069 | 0.6985 | |
| TCGA | PIAS4 | 51588 | RNAseq | 0.0913 | 0.1173 |
Upregulated datasets: 1; Downregulated datasets: 0.
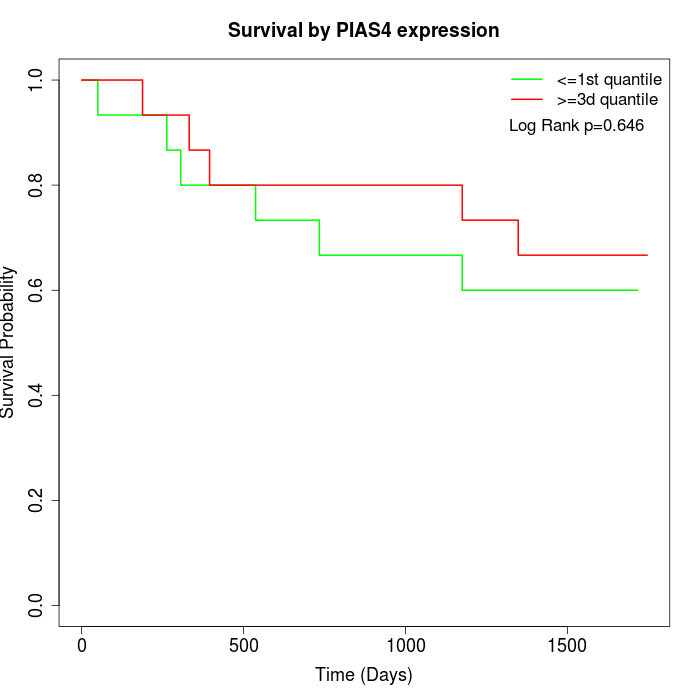
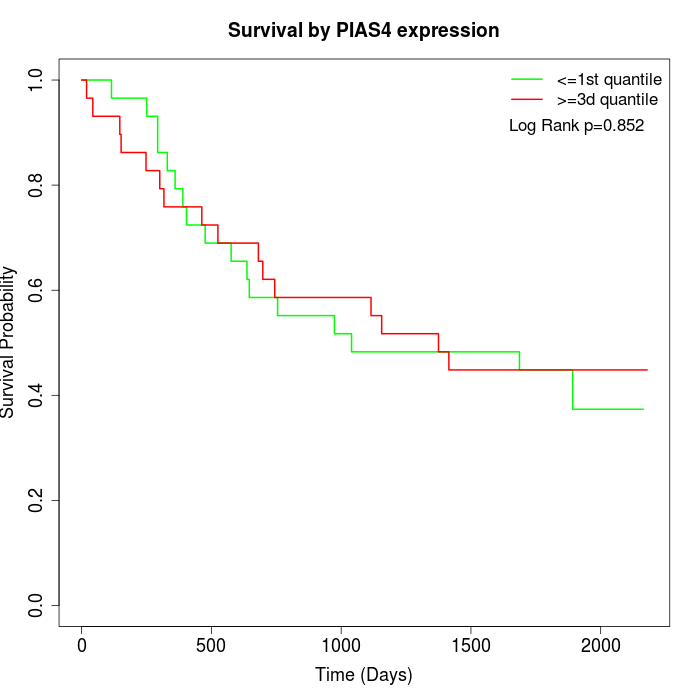
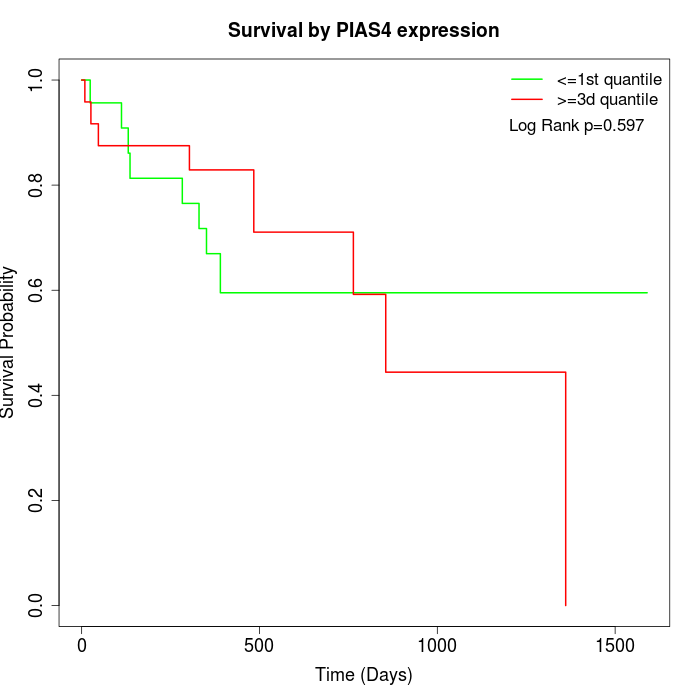
Survival by PIAS4 expression:
Note: Click image to view full size file.
Copy number change of PIAS4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PIAS4 | 51588 | 5 | 3 | 22 | |
| GSE20123 | PIAS4 | 51588 | 4 | 2 | 24 | |
| GSE43470 | PIAS4 | 51588 | 1 | 9 | 33 | |
| GSE46452 | PIAS4 | 51588 | 47 | 1 | 11 | |
| GSE47630 | PIAS4 | 51588 | 5 | 7 | 28 | |
| GSE54993 | PIAS4 | 51588 | 16 | 3 | 51 | |
| GSE54994 | PIAS4 | 51588 | 7 | 16 | 30 | |
| GSE60625 | PIAS4 | 51588 | 9 | 0 | 2 | |
| GSE74703 | PIAS4 | 51588 | 1 | 6 | 29 | |
| GSE74704 | PIAS4 | 51588 | 1 | 2 | 17 | |
| TCGA | PIAS4 | 51588 | 11 | 18 | 67 |
Total number of gains: 107; Total number of losses: 67; Total Number of normals: 314.
Somatic mutations of PIAS4:
Generating mutation plots.
Highly correlated genes for PIAS4:
Showing top 20/1465 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PIAS4 | NPHP3 | 0.794485 | 3 | 0 | 3 |
| PIAS4 | TMEM135 | 0.783677 | 3 | 0 | 3 |
| PIAS4 | MZT1 | 0.765576 | 3 | 0 | 3 |
| PIAS4 | ALKBH6 | 0.764 | 5 | 0 | 5 |
| PIAS4 | TMEM63B | 0.747871 | 4 | 0 | 4 |
| PIAS4 | WDR81 | 0.744871 | 4 | 0 | 4 |
| PIAS4 | BCL9L | 0.741582 | 3 | 0 | 3 |
| PIAS4 | SPIRE1 | 0.741108 | 3 | 0 | 3 |
| PIAS4 | ZNF286A | 0.73968 | 3 | 0 | 3 |
| PIAS4 | CPSF3 | 0.732364 | 4 | 0 | 3 |
| PIAS4 | SLC5A3 | 0.732225 | 4 | 0 | 3 |
| PIAS4 | MAK16 | 0.729898 | 4 | 0 | 4 |
| PIAS4 | COMMD2 | 0.721472 | 3 | 0 | 3 |
| PIAS4 | NCAPG | 0.717098 | 5 | 0 | 5 |
| PIAS4 | C1orf50 | 0.716072 | 3 | 0 | 3 |
| PIAS4 | RNASEH2B | 0.713066 | 3 | 0 | 3 |
| PIAS4 | LRRC58 | 0.712578 | 5 | 0 | 5 |
| PIAS4 | DGCR8 | 0.712308 | 3 | 0 | 3 |
| PIAS4 | SLC25A38 | 0.711319 | 3 | 0 | 3 |
| PIAS4 | AHCTF1 | 0.709713 | 6 | 0 | 6 |
For details and further investigation, click here