| Full name: DGCR8 microprocessor complex subunit | Alias Symbol: DGCRK6|Gy1|pasha | ||
| Type: protein-coding gene | Cytoband: 22q11.21 | ||
| Entrez ID: 54487 | HGNC ID: HGNC:2847 | Ensembl Gene: ENSG00000128191 | OMIM ID: 609030 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of DGCR8:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | DGCR8 | 54487 | 86304 | 0.3314 | 0.0000 | |
| GSE53624 | DGCR8 | 54487 | 86304 | 0.4821 | 0.0000 | |
| GSE97050 | DGCR8 | 54487 | A_23_P211355 | -0.0284 | 0.8994 | |
| SRP007169 | DGCR8 | 54487 | RNAseq | 0.8584 | 0.0450 | |
| SRP008496 | DGCR8 | 54487 | RNAseq | 1.0793 | 0.0006 | |
| SRP064894 | DGCR8 | 54487 | RNAseq | 0.4763 | 0.0174 | |
| SRP133303 | DGCR8 | 54487 | RNAseq | 0.1521 | 0.3345 | |
| SRP159526 | DGCR8 | 54487 | RNAseq | 0.3896 | 0.2796 | |
| SRP193095 | DGCR8 | 54487 | RNAseq | 0.3761 | 0.0001 | |
| SRP219564 | DGCR8 | 54487 | RNAseq | 0.1501 | 0.5473 | |
| TCGA | DGCR8 | 54487 | RNAseq | -0.0413 | 0.4014 |
Upregulated datasets: 1; Downregulated datasets: 0.
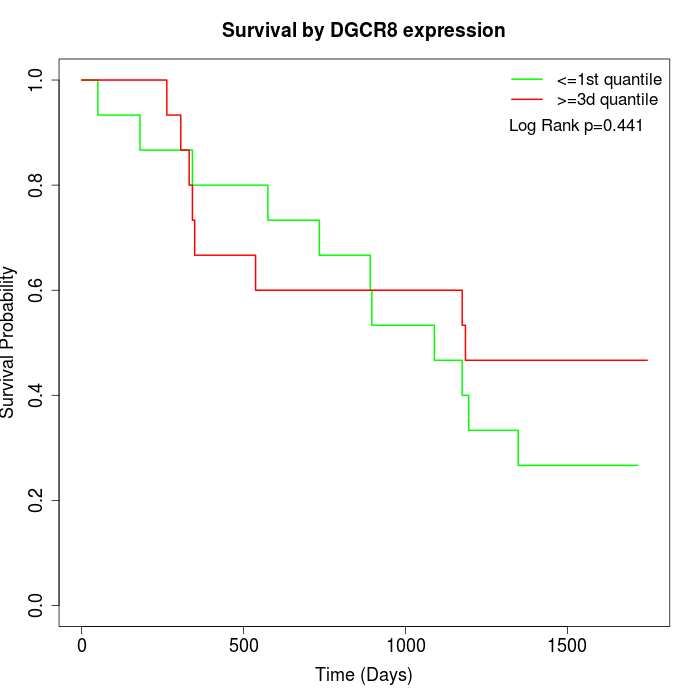
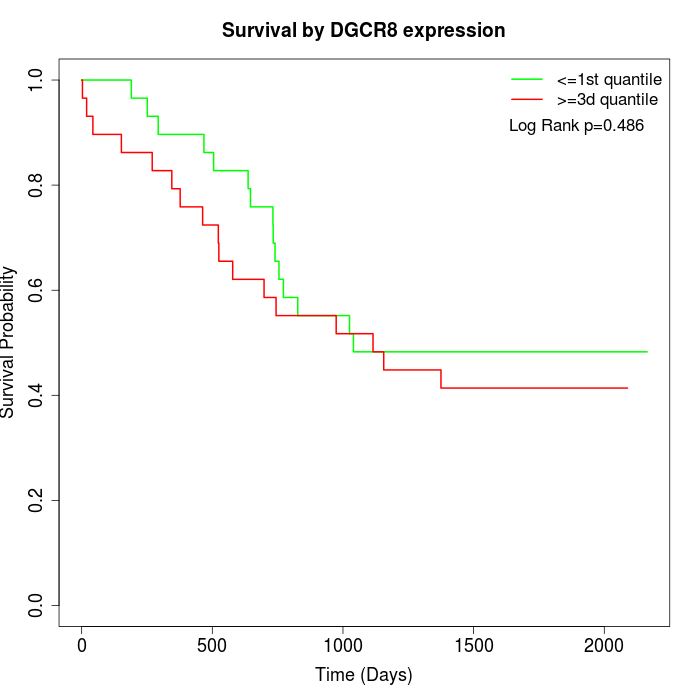
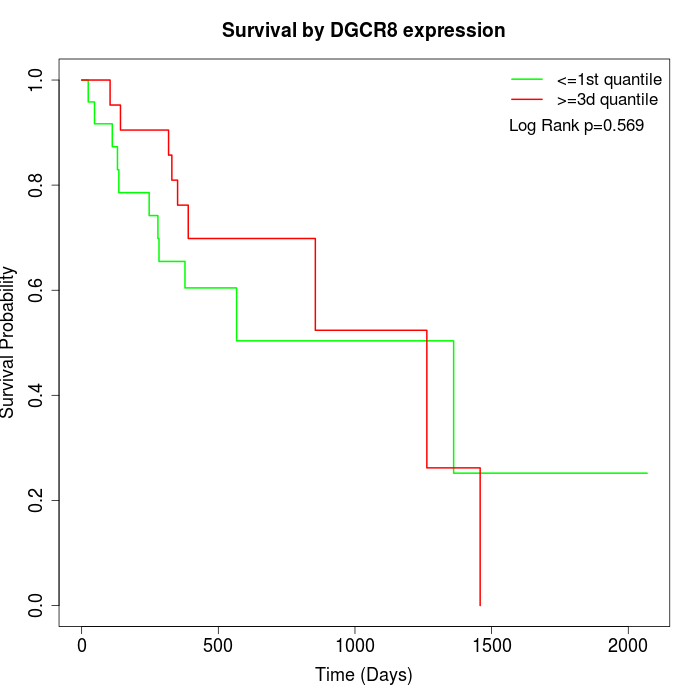
Survival by DGCR8 expression:
Note: Click image to view full size file.
Copy number change of DGCR8:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DGCR8 | 54487 | 5 | 7 | 18 | |
| GSE20123 | DGCR8 | 54487 | 5 | 7 | 18 | |
| GSE43470 | DGCR8 | 54487 | 4 | 7 | 32 | |
| GSE46452 | DGCR8 | 54487 | 33 | 1 | 25 | |
| GSE47630 | DGCR8 | 54487 | 8 | 5 | 27 | |
| GSE54993 | DGCR8 | 54487 | 3 | 6 | 61 | |
| GSE54994 | DGCR8 | 54487 | 12 | 7 | 34 | |
| GSE60625 | DGCR8 | 54487 | 5 | 0 | 6 | |
| GSE74703 | DGCR8 | 54487 | 4 | 5 | 27 | |
| GSE74704 | DGCR8 | 54487 | 3 | 4 | 13 | |
| TCGA | DGCR8 | 54487 | 27 | 17 | 52 |
Total number of gains: 109; Total number of losses: 66; Total Number of normals: 313.
Somatic mutations of DGCR8:
Generating mutation plots.
Highly correlated genes for DGCR8:
Showing top 20/127 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DGCR8 | SRRT | 0.794543 | 3 | 0 | 3 |
| DGCR8 | SENP3 | 0.778322 | 3 | 0 | 3 |
| DGCR8 | PRRC2A | 0.765236 | 3 | 0 | 3 |
| DGCR8 | MAP3K11 | 0.746197 | 3 | 0 | 3 |
| DGCR8 | SP3 | 0.738696 | 3 | 0 | 3 |
| DGCR8 | GMEB2 | 0.736064 | 3 | 0 | 3 |
| DGCR8 | RNPS1 | 0.733588 | 3 | 0 | 3 |
| DGCR8 | USP42 | 0.729998 | 3 | 0 | 3 |
| DGCR8 | ILF3 | 0.724765 | 4 | 0 | 4 |
| DGCR8 | SETD1A | 0.723598 | 4 | 0 | 4 |
| DGCR8 | MAZ | 0.723126 | 3 | 0 | 3 |
| DGCR8 | BRCA1 | 0.720983 | 3 | 0 | 3 |
| DGCR8 | POU2F1 | 0.720393 | 4 | 0 | 3 |
| DGCR8 | SF3A2 | 0.717317 | 4 | 0 | 4 |
| DGCR8 | TNPO3 | 0.716955 | 3 | 0 | 3 |
| DGCR8 | PIAS4 | 0.712308 | 3 | 0 | 3 |
| DGCR8 | SAMD1 | 0.712293 | 3 | 0 | 3 |
| DGCR8 | NVL | 0.711005 | 3 | 0 | 3 |
| DGCR8 | LSM2 | 0.71094 | 3 | 0 | 3 |
| DGCR8 | LMNB2 | 0.708357 | 3 | 0 | 3 |
For details and further investigation, click here