| Full name: phosphatidylinositol glycan anchor biosynthesis class O | Alias Symbol: DKFZp434M222|FLJ00135 | ||
| Type: protein-coding gene | Cytoband: 9p13.3 | ||
| Entrez ID: 84720 | HGNC ID: HGNC:23215 | Ensembl Gene: ENSG00000165282 | OMIM ID: 614730 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of PIGO:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PIGO | 84720 | 209998_at | -0.0134 | 0.9826 | |
| GSE20347 | PIGO | 84720 | 209998_at | 0.1318 | 0.2758 | |
| GSE23400 | PIGO | 84720 | 214990_at | -0.0941 | 0.0083 | |
| GSE26886 | PIGO | 84720 | 209998_at | -0.5021 | 0.0341 | |
| GSE29001 | PIGO | 84720 | 209998_at | 0.2995 | 0.1738 | |
| GSE38129 | PIGO | 84720 | 209998_at | 0.3502 | 0.0019 | |
| GSE45670 | PIGO | 84720 | 209998_at | 0.4632 | 0.0031 | |
| GSE63941 | PIGO | 84720 | 209998_at | -0.2287 | 0.6222 | |
| GSE77861 | PIGO | 84720 | 214990_at | -0.0287 | 0.8255 | |
| GSE97050 | PIGO | 84720 | A_23_P60240 | 0.1089 | 0.7991 | |
| SRP007169 | PIGO | 84720 | RNAseq | -0.0142 | 0.9745 | |
| SRP008496 | PIGO | 84720 | RNAseq | 0.2915 | 0.2562 | |
| SRP064894 | PIGO | 84720 | RNAseq | 0.6098 | 0.0000 | |
| SRP133303 | PIGO | 84720 | RNAseq | -0.0259 | 0.9011 | |
| SRP159526 | PIGO | 84720 | RNAseq | -0.0109 | 0.9578 | |
| SRP193095 | PIGO | 84720 | RNAseq | 0.0559 | 0.6536 | |
| SRP219564 | PIGO | 84720 | RNAseq | 0.7108 | 0.0605 | |
| TCGA | PIGO | 84720 | RNAseq | -0.0246 | 0.6566 |
Upregulated datasets: 0; Downregulated datasets: 0.
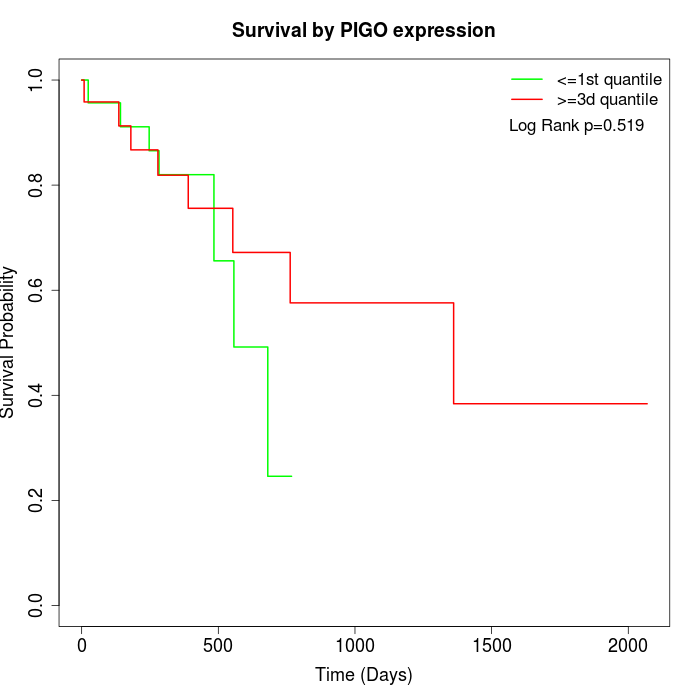
Survival by PIGO expression:
Note: Click image to view full size file.
Copy number change of PIGO:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PIGO | 84720 | 3 | 13 | 14 | |
| GSE20123 | PIGO | 84720 | 3 | 13 | 14 | |
| GSE43470 | PIGO | 84720 | 3 | 10 | 30 | |
| GSE46452 | PIGO | 84720 | 6 | 15 | 38 | |
| GSE47630 | PIGO | 84720 | 1 | 20 | 19 | |
| GSE54993 | PIGO | 84720 | 6 | 0 | 64 | |
| GSE54994 | PIGO | 84720 | 6 | 12 | 35 | |
| GSE60625 | PIGO | 84720 | 0 | 0 | 11 | |
| GSE74703 | PIGO | 84720 | 2 | 7 | 27 | |
| GSE74704 | PIGO | 84720 | 0 | 11 | 9 | |
| TCGA | PIGO | 84720 | 16 | 44 | 36 |
Total number of gains: 46; Total number of losses: 145; Total Number of normals: 297.
Somatic mutations of PIGO:
Generating mutation plots.
Highly correlated genes for PIGO:
Showing top 20/64 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PIGO | STOML2 | 0.662568 | 7 | 0 | 7 |
| PIGO | ASIC1 | 0.641883 | 3 | 0 | 3 |
| PIGO | WWP2 | 0.63574 | 4 | 0 | 3 |
| PIGO | TOMM5 | 0.628317 | 3 | 0 | 3 |
| PIGO | NEU1 | 0.615302 | 3 | 0 | 3 |
| PIGO | PHACTR1 | 0.614368 | 3 | 0 | 3 |
| PIGO | CYP2F1 | 0.612269 | 3 | 0 | 3 |
| PIGO | EPHB2 | 0.602735 | 4 | 0 | 4 |
| PIGO | RNF170 | 0.597189 | 3 | 0 | 3 |
| PIGO | ATP6V0B | 0.590465 | 4 | 0 | 4 |
| PIGO | S100Z | 0.589957 | 3 | 0 | 3 |
| PIGO | CYB561D2 | 0.583077 | 3 | 0 | 3 |
| PIGO | TDP1 | 0.579945 | 5 | 0 | 5 |
| PIGO | TPX2 | 0.565772 | 4 | 0 | 4 |
| PIGO | SLC25A5 | 0.563852 | 4 | 0 | 3 |
| PIGO | HDLBP | 0.562692 | 3 | 0 | 3 |
| PIGO | MRPS12 | 0.557753 | 5 | 0 | 4 |
| PIGO | CLPB | 0.556822 | 6 | 0 | 4 |
| PIGO | NXT2 | 0.555527 | 4 | 0 | 3 |
| PIGO | E2F2 | 0.554171 | 3 | 0 | 3 |
For details and further investigation, click here