| Full name: ATPase H+ transporting V0 subunit b | Alias Symbol: VMA16|HATPL | ||
| Type: protein-coding gene | Cytoband: 1p34.1 | ||
| Entrez ID: 533 | HGNC ID: HGNC:861 | Ensembl Gene: ENSG00000117410 | OMIM ID: 603717 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
ATP6V0B involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa05110 | Vibrio cholerae infection | |
| hsa05120 | Epithelial cell signaling in Helicobacter pylori infection | |
| hsa05152 | Tuberculosis |
Expression of ATP6V0B:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ATP6V0B | 533 | 200078_s_at | 0.6064 | 0.2519 | |
| GSE20347 | ATP6V0B | 533 | 200078_s_at | 0.3746 | 0.0124 | |
| GSE23400 | ATP6V0B | 533 | 200078_s_at | 0.4653 | 0.0000 | |
| GSE26886 | ATP6V0B | 533 | 200078_s_at | 0.0609 | 0.8320 | |
| GSE29001 | ATP6V0B | 533 | 200078_s_at | 0.1008 | 0.7986 | |
| GSE38129 | ATP6V0B | 533 | 200078_s_at | 0.6153 | 0.0000 | |
| GSE45670 | ATP6V0B | 533 | 200078_s_at | 0.5723 | 0.0004 | |
| GSE53622 | ATP6V0B | 533 | 6546 | 0.3234 | 0.0000 | |
| GSE53624 | ATP6V0B | 533 | 6546 | 0.5806 | 0.0000 | |
| GSE63941 | ATP6V0B | 533 | 200078_s_at | 1.2469 | 0.0098 | |
| GSE77861 | ATP6V0B | 533 | 200078_s_at | 0.2557 | 0.4592 | |
| GSE97050 | ATP6V0B | 533 | A_23_P137814 | 0.4351 | 0.1581 | |
| SRP007169 | ATP6V0B | 533 | RNAseq | -0.6602 | 0.1116 | |
| SRP008496 | ATP6V0B | 533 | RNAseq | -0.6677 | 0.0019 | |
| SRP064894 | ATP6V0B | 533 | RNAseq | 0.8368 | 0.0030 | |
| SRP133303 | ATP6V0B | 533 | RNAseq | 0.6902 | 0.0000 | |
| SRP159526 | ATP6V0B | 533 | RNAseq | 0.5546 | 0.0099 | |
| SRP193095 | ATP6V0B | 533 | RNAseq | 0.2458 | 0.0824 | |
| SRP219564 | ATP6V0B | 533 | RNAseq | 0.7531 | 0.0701 | |
| TCGA | ATP6V0B | 533 | RNAseq | -0.0268 | 0.5809 |
Upregulated datasets: 1; Downregulated datasets: 0.
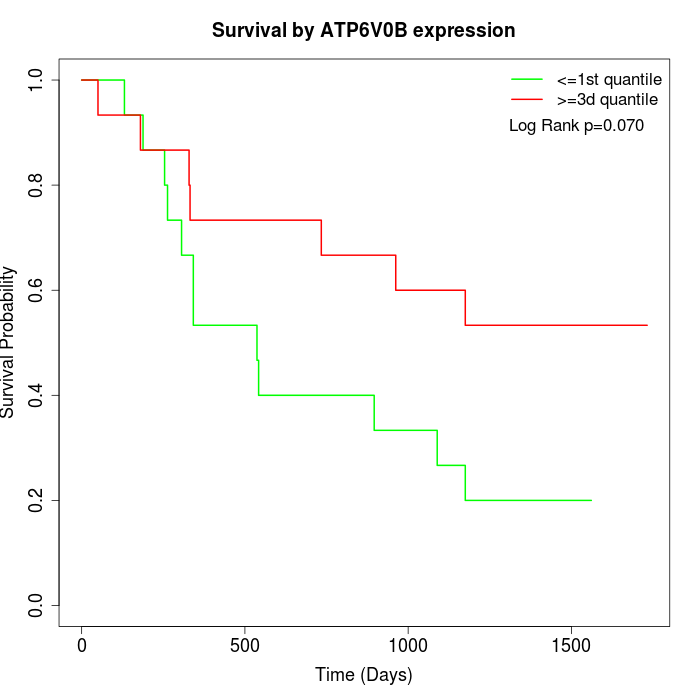
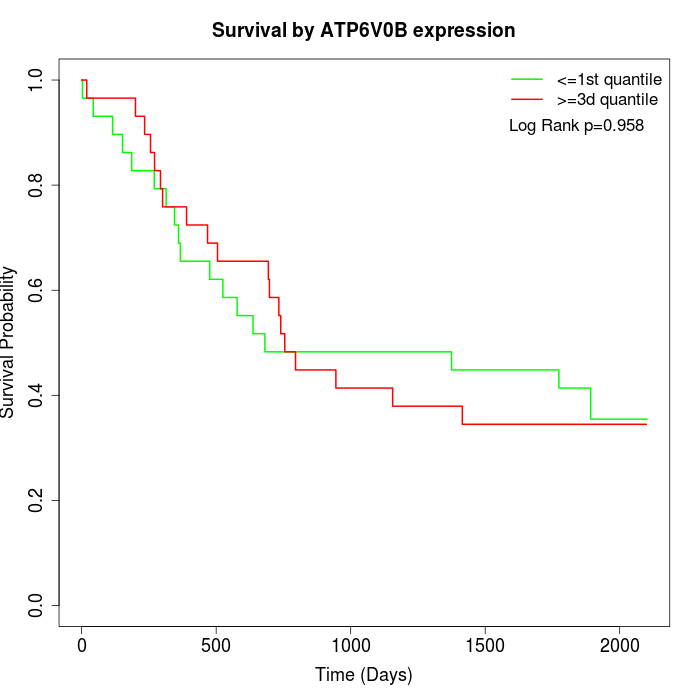
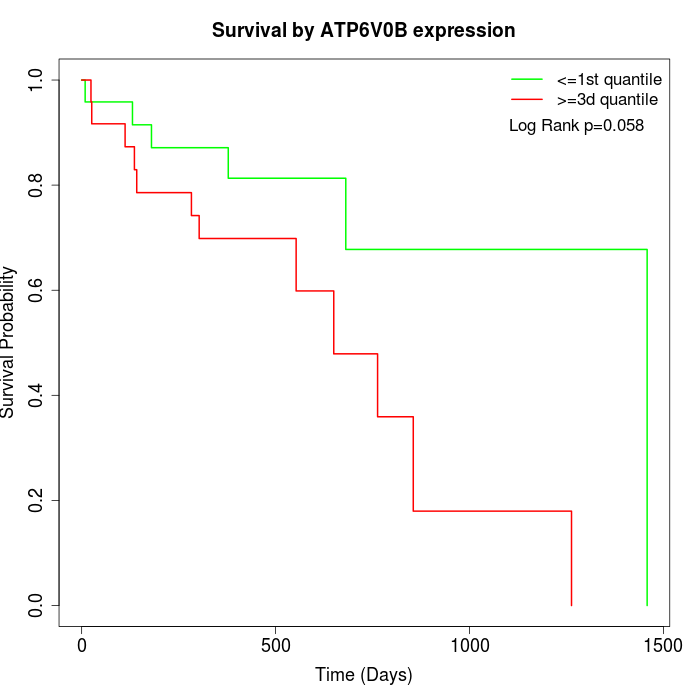
Survival by ATP6V0B expression:
Note: Click image to view full size file.
Copy number change of ATP6V0B:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ATP6V0B | 533 | 4 | 3 | 23 | |
| GSE20123 | ATP6V0B | 533 | 4 | 2 | 24 | |
| GSE43470 | ATP6V0B | 533 | 7 | 2 | 34 | |
| GSE46452 | ATP6V0B | 533 | 2 | 1 | 56 | |
| GSE47630 | ATP6V0B | 533 | 9 | 3 | 28 | |
| GSE54993 | ATP6V0B | 533 | 0 | 2 | 68 | |
| GSE54994 | ATP6V0B | 533 | 14 | 2 | 37 | |
| GSE60625 | ATP6V0B | 533 | 0 | 0 | 11 | |
| GSE74703 | ATP6V0B | 533 | 6 | 1 | 29 | |
| GSE74704 | ATP6V0B | 533 | 2 | 0 | 18 | |
| TCGA | ATP6V0B | 533 | 14 | 16 | 66 |
Total number of gains: 62; Total number of losses: 32; Total Number of normals: 394.
Somatic mutations of ATP6V0B:
Generating mutation plots.
Highly correlated genes for ATP6V0B:
Showing top 20/1294 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ATP6V0B | INTS9 | 0.831149 | 3 | 0 | 3 |
| ATP6V0B | SPNS1 | 0.815595 | 3 | 0 | 3 |
| ATP6V0B | DUS3L | 0.791376 | 3 | 0 | 3 |
| ATP6V0B | KTI12 | 0.780422 | 3 | 0 | 3 |
| ATP6V0B | POLDIP2 | 0.77629 | 3 | 0 | 3 |
| ATP6V0B | FBXW8 | 0.751016 | 3 | 0 | 3 |
| ATP6V0B | STUB1 | 0.740557 | 3 | 0 | 3 |
| ATP6V0B | RRP36 | 0.729891 | 5 | 0 | 5 |
| ATP6V0B | TOMM22 | 0.724618 | 5 | 0 | 4 |
| ATP6V0B | FAM89A | 0.720396 | 3 | 0 | 3 |
| ATP6V0B | ISY1 | 0.720338 | 3 | 0 | 3 |
| ATP6V0B | FASTKD2 | 0.719229 | 4 | 0 | 4 |
| ATP6V0B | THRAP3 | 0.716886 | 3 | 0 | 3 |
| ATP6V0B | PPIL1 | 0.713736 | 5 | 0 | 4 |
| ATP6V0B | TM9SF3 | 0.71189 | 3 | 0 | 3 |
| ATP6V0B | SNX17 | 0.709771 | 3 | 0 | 3 |
| ATP6V0B | ALG10 | 0.697265 | 4 | 0 | 3 |
| ATP6V0B | METTL23 | 0.696754 | 4 | 0 | 3 |
| ATP6V0B | DDOST | 0.696684 | 10 | 0 | 10 |
| ATP6V0B | CIB1 | 0.695025 | 4 | 0 | 3 |
For details and further investigation, click here