| Full name: phosphatidylinositol glycan anchor biosynthesis class Z | Alias Symbol: FLJ12768|MGC52163|SMP3 | ||
| Type: protein-coding gene | Cytoband: 3q29 | ||
| Entrez ID: 80235 | HGNC ID: HGNC:30596 | Ensembl Gene: ENSG00000119227 | OMIM ID: 611671 |
| Drug and gene relationship at DGIdb | |||
Expression of PIGZ:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PIGZ | 80235 | 220041_at | -0.0352 | 0.9676 | |
| GSE20347 | PIGZ | 80235 | 220041_at | 0.2603 | 0.1220 | |
| GSE23400 | PIGZ | 80235 | 220041_at | 0.1825 | 0.0022 | |
| GSE26886 | PIGZ | 80235 | 220041_at | 0.5343 | 0.0172 | |
| GSE29001 | PIGZ | 80235 | 220041_at | 0.4703 | 0.1016 | |
| GSE38129 | PIGZ | 80235 | 220041_at | 0.1415 | 0.4283 | |
| GSE45670 | PIGZ | 80235 | 220041_at | -0.1599 | 0.4082 | |
| GSE53622 | PIGZ | 80235 | 124239 | 0.0755 | 0.6151 | |
| GSE53624 | PIGZ | 80235 | 124239 | 0.3267 | 0.0013 | |
| GSE63941 | PIGZ | 80235 | 220041_at | -0.0507 | 0.9419 | |
| GSE77861 | PIGZ | 80235 | 220041_at | 0.3028 | 0.1936 | |
| GSE97050 | PIGZ | 80235 | A_23_P143935 | 0.0143 | 0.9705 | |
| SRP064894 | PIGZ | 80235 | RNAseq | 0.6230 | 0.0621 | |
| SRP133303 | PIGZ | 80235 | RNAseq | 0.0979 | 0.7009 | |
| SRP159526 | PIGZ | 80235 | RNAseq | 0.9227 | 0.0931 | |
| SRP193095 | PIGZ | 80235 | RNAseq | 0.6925 | 0.0005 | |
| SRP219564 | PIGZ | 80235 | RNAseq | 0.7000 | 0.1697 | |
| TCGA | PIGZ | 80235 | RNAseq | 0.0328 | 0.7687 |
Upregulated datasets: 0; Downregulated datasets: 0.
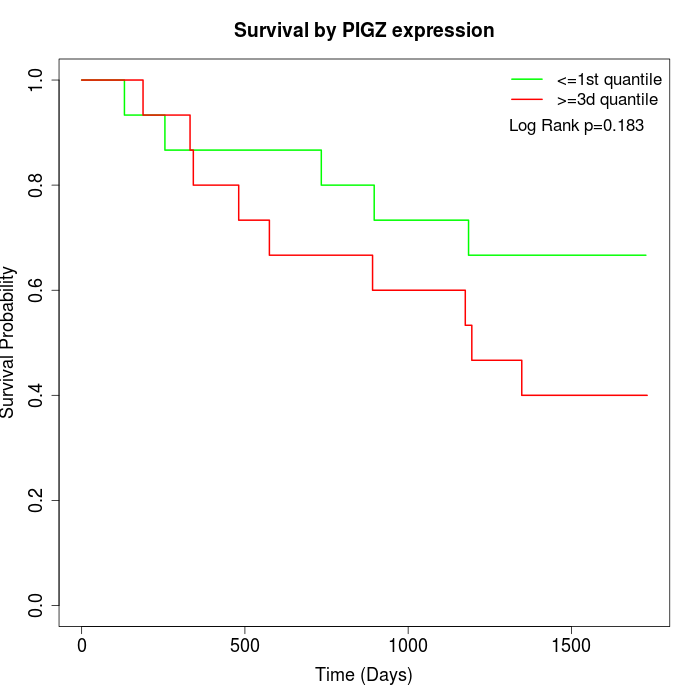
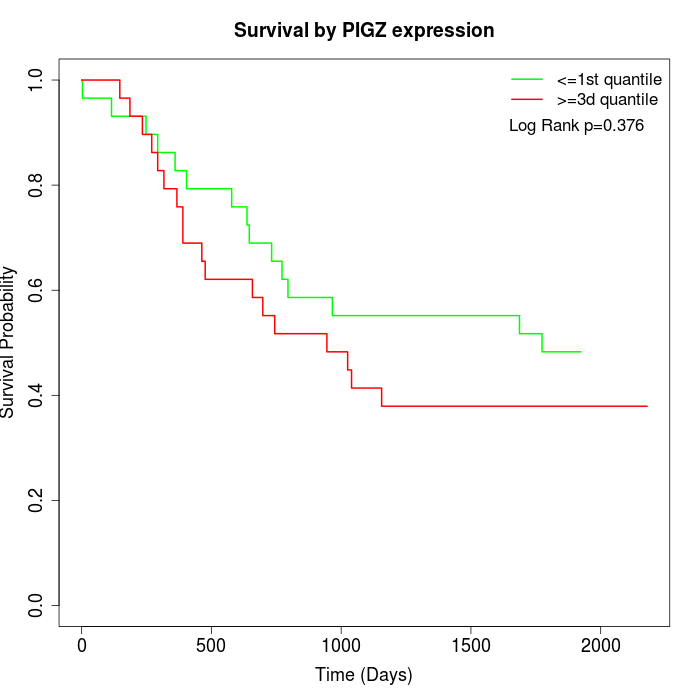
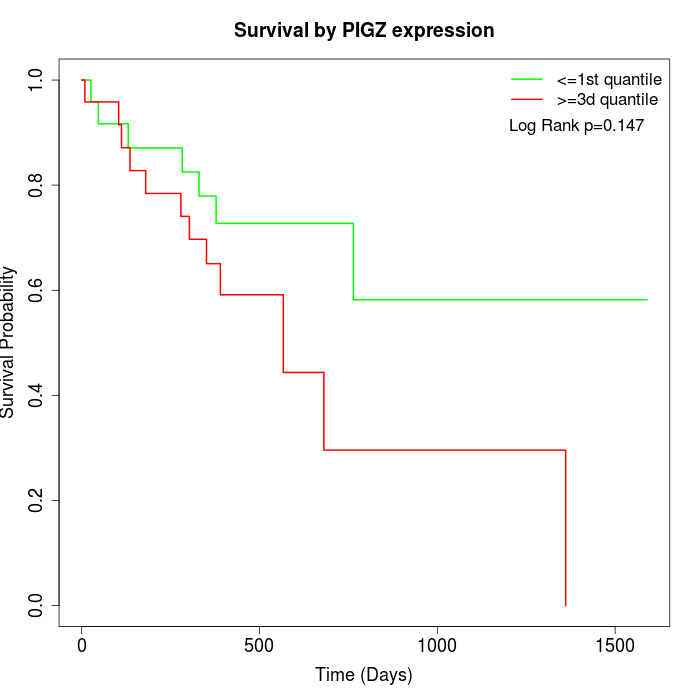
Survival by PIGZ expression:
Note: Click image to view full size file.
Copy number change of PIGZ:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PIGZ | 80235 | 22 | 0 | 8 | |
| GSE20123 | PIGZ | 80235 | 22 | 0 | 8 | |
| GSE43470 | PIGZ | 80235 | 21 | 0 | 22 | |
| GSE46452 | PIGZ | 80235 | 20 | 1 | 38 | |
| GSE47630 | PIGZ | 80235 | 27 | 2 | 11 | |
| GSE54993 | PIGZ | 80235 | 1 | 22 | 47 | |
| GSE54994 | PIGZ | 80235 | 41 | 0 | 12 | |
| GSE60625 | PIGZ | 80235 | 0 | 6 | 5 | |
| GSE74703 | PIGZ | 80235 | 19 | 0 | 17 | |
| GSE74704 | PIGZ | 80235 | 14 | 0 | 6 | |
| TCGA | PIGZ | 80235 | 76 | 1 | 19 |
Total number of gains: 263; Total number of losses: 32; Total Number of normals: 193.
Somatic mutations of PIGZ:
Generating mutation plots.
Highly correlated genes for PIGZ:
Showing top 20/82 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PIGZ | CNTNAP2 | 0.663284 | 3 | 0 | 3 |
| PIGZ | KIF18B | 0.660037 | 3 | 0 | 3 |
| PIGZ | ENOSF1 | 0.646279 | 3 | 0 | 3 |
| PIGZ | MCM8 | 0.641284 | 3 | 0 | 3 |
| PIGZ | TMEM223 | 0.622568 | 3 | 0 | 3 |
| PIGZ | PPP6R3 | 0.61909 | 3 | 0 | 3 |
| PIGZ | SLC36A1 | 0.614732 | 3 | 0 | 3 |
| PIGZ | FJX1 | 0.613759 | 3 | 0 | 3 |
| PIGZ | HHIPL2 | 0.610105 | 3 | 0 | 3 |
| PIGZ | ARV1 | 0.609889 | 3 | 0 | 3 |
| PIGZ | DMRTC2 | 0.606596 | 3 | 0 | 3 |
| PIGZ | S100A6 | 0.600477 | 4 | 0 | 3 |
| PIGZ | SNCAIP | 0.597232 | 4 | 0 | 3 |
| PIGZ | FAM160A2 | 0.594675 | 4 | 0 | 3 |
| PIGZ | VAMP2 | 0.594655 | 4 | 0 | 3 |
| PIGZ | TRPC1 | 0.588004 | 5 | 0 | 4 |
| PIGZ | CTBP1 | 0.587135 | 3 | 0 | 3 |
| PIGZ | GTSE1 | 0.584958 | 3 | 0 | 3 |
| PIGZ | PRR5L | 0.579356 | 4 | 0 | 4 |
| PIGZ | NIPSNAP1 | 0.578504 | 3 | 0 | 3 |
For details and further investigation, click here