| Full name: phosphoinositide kinase, FYVE-type zinc finger containing | Alias Symbol: MGC40423|KIAA0981|PIKfyve|PIP5K|p235|ZFYVE29|FAB1 | ||
| Type: protein-coding gene | Cytoband: 2q34 | ||
| Entrez ID: 200576 | HGNC ID: HGNC:23785 | Ensembl Gene: ENSG00000115020 | OMIM ID: 609414 |
| Drug and gene relationship at DGIdb | |||
PIKFYVE involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04810 | Regulation of actin cytoskeleton |
Expression of PIKFYVE:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PIKFYVE | 200576 | 213111_at | -0.1212 | 0.6765 | |
| GSE20347 | PIKFYVE | 200576 | 213111_at | -0.0195 | 0.9313 | |
| GSE23400 | PIKFYVE | 200576 | 213111_at | 0.1772 | 0.0398 | |
| GSE26886 | PIKFYVE | 200576 | 213111_at | -0.1368 | 0.7797 | |
| GSE29001 | PIKFYVE | 200576 | 213111_at | 0.1705 | 0.6985 | |
| GSE38129 | PIKFYVE | 200576 | 213111_at | -0.0317 | 0.8693 | |
| GSE45670 | PIKFYVE | 200576 | 213111_at | -0.1471 | 0.3585 | |
| GSE53622 | PIKFYVE | 200576 | 142084 | 0.2198 | 0.0032 | |
| GSE53624 | PIKFYVE | 200576 | 41840 | -0.1557 | 0.1675 | |
| GSE63941 | PIKFYVE | 200576 | 213111_at | -1.6611 | 0.0007 | |
| GSE77861 | PIKFYVE | 200576 | 213111_at | 0.5676 | 0.0514 | |
| GSE97050 | PIKFYVE | 200576 | A_33_P3348973 | 0.3442 | 0.2773 | |
| SRP007169 | PIKFYVE | 200576 | RNAseq | 0.6539 | 0.1127 | |
| SRP008496 | PIKFYVE | 200576 | RNAseq | 0.6845 | 0.0028 | |
| SRP064894 | PIKFYVE | 200576 | RNAseq | -0.0820 | 0.8054 | |
| SRP133303 | PIKFYVE | 200576 | RNAseq | 0.1434 | 0.1356 | |
| SRP159526 | PIKFYVE | 200576 | RNAseq | 0.0234 | 0.9311 | |
| SRP193095 | PIKFYVE | 200576 | RNAseq | 0.0411 | 0.6476 | |
| SRP219564 | PIKFYVE | 200576 | RNAseq | -0.2663 | 0.3898 | |
| TCGA | PIKFYVE | 200576 | RNAseq | -0.1105 | 0.0424 |
Upregulated datasets: 0; Downregulated datasets: 1.
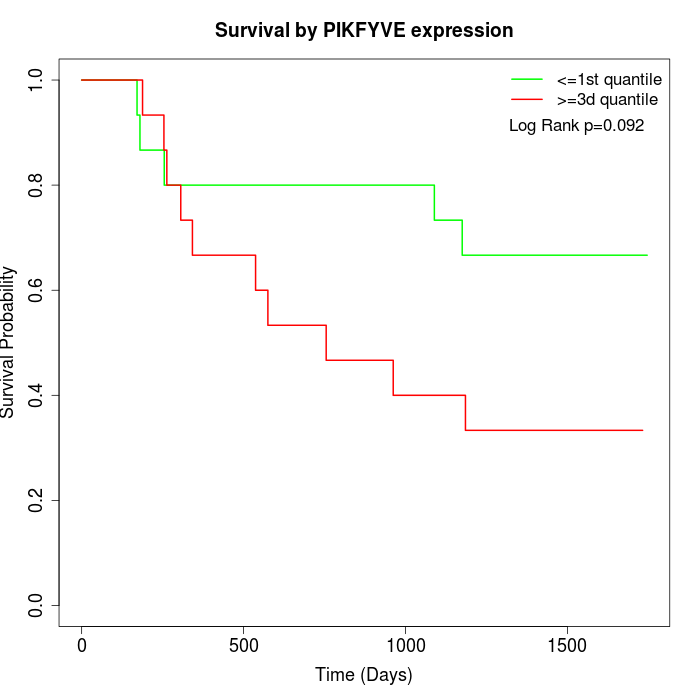
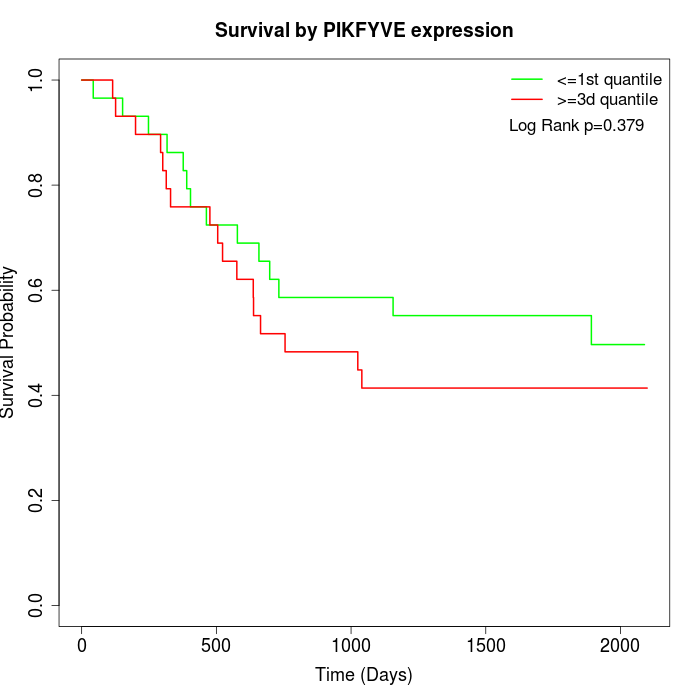
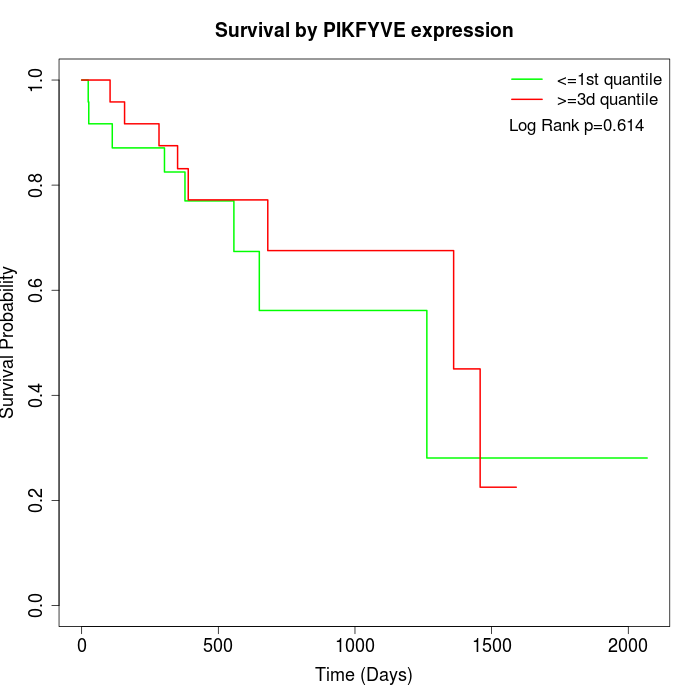
Survival by PIKFYVE expression:
Note: Click image to view full size file.
Copy number change of PIKFYVE:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PIKFYVE | 200576 | 3 | 10 | 17 | |
| GSE20123 | PIKFYVE | 200576 | 3 | 10 | 17 | |
| GSE43470 | PIKFYVE | 200576 | 2 | 4 | 37 | |
| GSE46452 | PIKFYVE | 200576 | 0 | 5 | 54 | |
| GSE47630 | PIKFYVE | 200576 | 4 | 5 | 31 | |
| GSE54993 | PIKFYVE | 200576 | 0 | 3 | 67 | |
| GSE54994 | PIKFYVE | 200576 | 9 | 9 | 35 | |
| GSE60625 | PIKFYVE | 200576 | 0 | 3 | 8 | |
| GSE74703 | PIKFYVE | 200576 | 2 | 3 | 31 | |
| GSE74704 | PIKFYVE | 200576 | 2 | 5 | 13 | |
| TCGA | PIKFYVE | 200576 | 15 | 20 | 61 |
Total number of gains: 40; Total number of losses: 77; Total Number of normals: 371.
Somatic mutations of PIKFYVE:
Generating mutation plots.
Highly correlated genes for PIKFYVE:
Showing top 20/423 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PIKFYVE | MYSM1 | 0.742898 | 3 | 0 | 3 |
| PIKFYVE | TTL | 0.708578 | 3 | 0 | 3 |
| PIKFYVE | RELL1 | 0.702304 | 3 | 0 | 3 |
| PIKFYVE | C12orf66 | 0.701324 | 4 | 0 | 4 |
| PIKFYVE | JOSD1 | 0.695306 | 3 | 0 | 3 |
| PIKFYVE | GLRX3 | 0.694977 | 3 | 0 | 3 |
| PIKFYVE | CDH13 | 0.694502 | 3 | 0 | 3 |
| PIKFYVE | UBE4A | 0.689086 | 3 | 0 | 3 |
| PIKFYVE | RB1 | 0.686725 | 5 | 0 | 5 |
| PIKFYVE | RAB8B | 0.685724 | 3 | 0 | 3 |
| PIKFYVE | SLC22A15 | 0.684296 | 4 | 0 | 3 |
| PIKFYVE | SLC44A1 | 0.683073 | 3 | 0 | 3 |
| PIKFYVE | BCS1L | 0.680952 | 3 | 0 | 3 |
| PIKFYVE | AMIGO2 | 0.679571 | 3 | 0 | 3 |
| PIKFYVE | RPAP3 | 0.67795 | 4 | 0 | 3 |
| PIKFYVE | GLS | 0.677746 | 3 | 0 | 3 |
| PIKFYVE | ACBD5 | 0.675089 | 3 | 0 | 3 |
| PIKFYVE | C12orf49 | 0.674351 | 3 | 0 | 3 |
| PIKFYVE | BLOC1S6 | 0.674047 | 3 | 0 | 3 |
| PIKFYVE | KIAA0100 | 0.673211 | 3 | 0 | 3 |
For details and further investigation, click here