| Full name: phosphatidylinositol-5-phosphate 4-kinase type 2 gamma | Alias Symbol: FLJ22055 | ||
| Type: protein-coding gene | Cytoband: 12q13.3 | ||
| Entrez ID: 79837 | HGNC ID: HGNC:23786 | Ensembl Gene: ENSG00000166908 | OMIM ID: 617104 |
| Drug and gene relationship at DGIdb | |||
PIP4K2C involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04810 | Regulation of actin cytoskeleton |
Expression of PIP4K2C:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PIP4K2C | 79837 | 218942_at | 0.3067 | 0.3178 | |
| GSE20347 | PIP4K2C | 79837 | 218942_at | 0.0115 | 0.9088 | |
| GSE23400 | PIP4K2C | 79837 | 218942_at | 0.2546 | 0.0000 | |
| GSE26886 | PIP4K2C | 79837 | 218942_at | 0.2140 | 0.3325 | |
| GSE29001 | PIP4K2C | 79837 | 218942_at | 0.0806 | 0.7292 | |
| GSE38129 | PIP4K2C | 79837 | 218942_at | 0.3916 | 0.0034 | |
| GSE45670 | PIP4K2C | 79837 | 218942_at | 0.3722 | 0.0003 | |
| GSE53622 | PIP4K2C | 79837 | 41437 | 0.4946 | 0.0000 | |
| GSE53624 | PIP4K2C | 79837 | 41437 | 0.3825 | 0.0000 | |
| GSE63941 | PIP4K2C | 79837 | 218942_at | 0.5914 | 0.1127 | |
| GSE77861 | PIP4K2C | 79837 | 218942_at | 0.4225 | 0.0335 | |
| GSE97050 | PIP4K2C | 79837 | A_23_P162199 | 0.7179 | 0.1592 | |
| SRP007169 | PIP4K2C | 79837 | RNAseq | -0.2469 | 0.4738 | |
| SRP008496 | PIP4K2C | 79837 | RNAseq | -0.4496 | 0.0155 | |
| SRP064894 | PIP4K2C | 79837 | RNAseq | 0.2697 | 0.1822 | |
| SRP133303 | PIP4K2C | 79837 | RNAseq | 0.6729 | 0.0000 | |
| SRP159526 | PIP4K2C | 79837 | RNAseq | 0.1138 | 0.4893 | |
| SRP193095 | PIP4K2C | 79837 | RNAseq | 0.0757 | 0.6231 | |
| SRP219564 | PIP4K2C | 79837 | RNAseq | 0.4020 | 0.2815 | |
| TCGA | PIP4K2C | 79837 | RNAseq | -0.0341 | 0.4726 |
Upregulated datasets: 0; Downregulated datasets: 0.
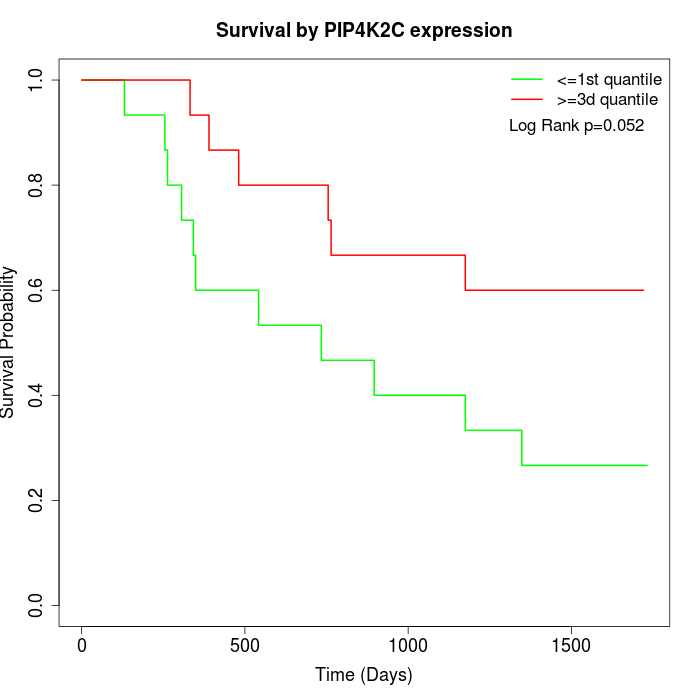
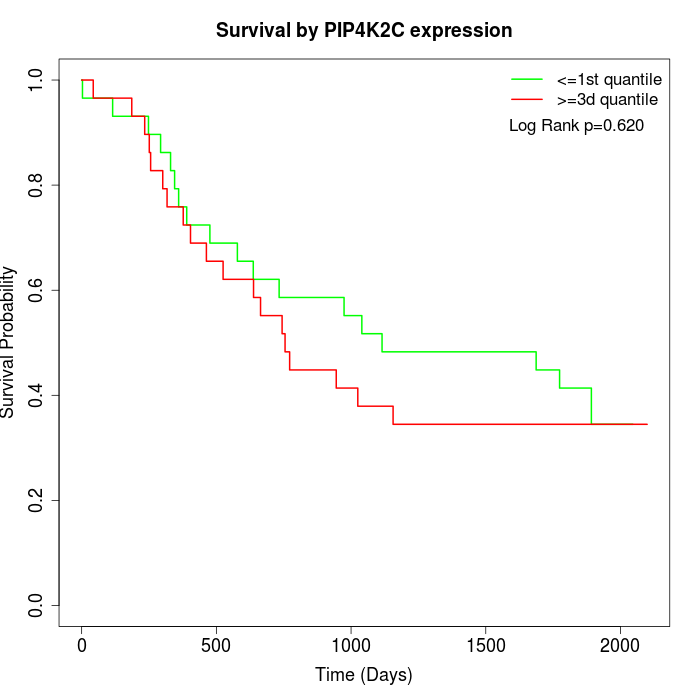
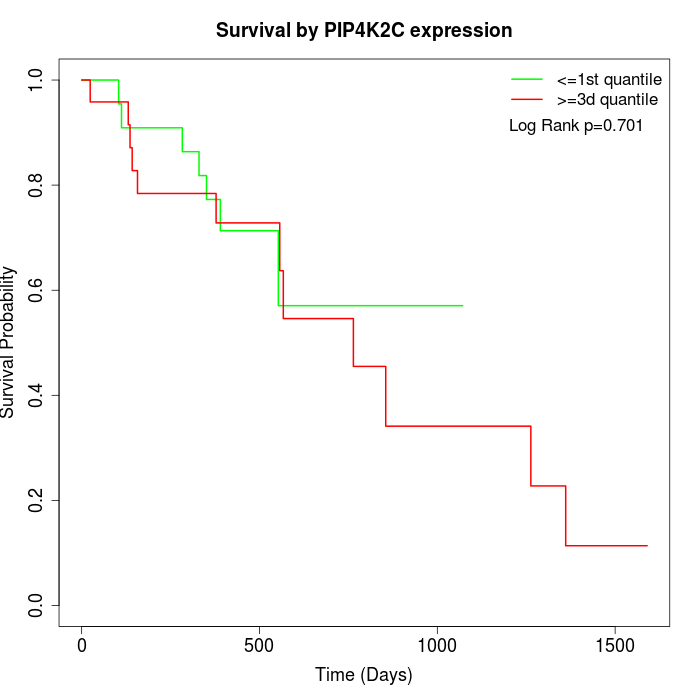
Survival by PIP4K2C expression:
Note: Click image to view full size file.
Copy number change of PIP4K2C:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PIP4K2C | 79837 | 7 | 1 | 22 | |
| GSE20123 | PIP4K2C | 79837 | 7 | 0 | 23 | |
| GSE43470 | PIP4K2C | 79837 | 2 | 0 | 41 | |
| GSE46452 | PIP4K2C | 79837 | 8 | 1 | 50 | |
| GSE47630 | PIP4K2C | 79837 | 10 | 2 | 28 | |
| GSE54993 | PIP4K2C | 79837 | 0 | 5 | 65 | |
| GSE54994 | PIP4K2C | 79837 | 4 | 1 | 48 | |
| GSE60625 | PIP4K2C | 79837 | 0 | 0 | 11 | |
| GSE74703 | PIP4K2C | 79837 | 2 | 0 | 34 | |
| GSE74704 | PIP4K2C | 79837 | 5 | 0 | 15 | |
| TCGA | PIP4K2C | 79837 | 14 | 10 | 72 |
Total number of gains: 59; Total number of losses: 20; Total Number of normals: 409.
Somatic mutations of PIP4K2C:
Generating mutation plots.
Highly correlated genes for PIP4K2C:
Showing top 20/1035 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PIP4K2C | FARS2 | 0.814045 | 3 | 0 | 3 |
| PIP4K2C | RAB1B | 0.809902 | 3 | 0 | 3 |
| PIP4K2C | SLC37A3 | 0.794793 | 3 | 0 | 3 |
| PIP4K2C | ZNF3 | 0.788748 | 3 | 0 | 3 |
| PIP4K2C | DDB1 | 0.781535 | 3 | 0 | 3 |
| PIP4K2C | THAP11 | 0.779814 | 3 | 0 | 3 |
| PIP4K2C | USP37 | 0.768443 | 3 | 0 | 3 |
| PIP4K2C | ARL13B | 0.761735 | 3 | 0 | 3 |
| PIP4K2C | NCOA6 | 0.759141 | 3 | 0 | 3 |
| PIP4K2C | ALG1 | 0.756191 | 4 | 0 | 3 |
| PIP4K2C | PI4K2A | 0.752436 | 3 | 0 | 3 |
| PIP4K2C | ZBTB9 | 0.747151 | 4 | 0 | 4 |
| PIP4K2C | LGALS8 | 0.739047 | 4 | 0 | 3 |
| PIP4K2C | NUP133 | 0.735528 | 3 | 0 | 3 |
| PIP4K2C | SFPQ | 0.734516 | 4 | 0 | 4 |
| PIP4K2C | RNF40 | 0.734154 | 4 | 0 | 4 |
| PIP4K2C | KRTCAP2 | 0.733878 | 3 | 0 | 3 |
| PIP4K2C | CHD8 | 0.730403 | 3 | 0 | 3 |
| PIP4K2C | TM9SF1 | 0.729231 | 3 | 0 | 3 |
| PIP4K2C | ALG10 | 0.723671 | 4 | 0 | 4 |
For details and further investigation, click here