| Full name: phospholipid scramblase 2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 3q24 | ||
| Entrez ID: 57047 | HGNC ID: HGNC:16494 | Ensembl Gene: ENSG00000163746 | OMIM ID: 607610 |
| Drug and gene relationship at DGIdb | |||
Expression of PLSCR2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PLSCR2 | 57047 | 207374_at | 0.2292 | 0.3384 | |
| GSE20347 | PLSCR2 | 57047 | 207374_at | 0.2780 | 0.0050 | |
| GSE23400 | PLSCR2 | 57047 | 207374_at | 0.0587 | 0.1926 | |
| GSE26886 | PLSCR2 | 57047 | 207374_at | 0.0176 | 0.8971 | |
| GSE29001 | PLSCR2 | 57047 | 207374_at | 0.4133 | 0.0297 | |
| GSE38129 | PLSCR2 | 57047 | 207374_at | 0.1831 | 0.0608 | |
| GSE45670 | PLSCR2 | 57047 | 207374_at | 0.3325 | 0.0014 | |
| GSE53622 | PLSCR2 | 57047 | 38465 | 0.2851 | 0.0234 | |
| GSE53624 | PLSCR2 | 57047 | 38465 | 0.3372 | 0.0113 | |
| GSE63941 | PLSCR2 | 57047 | 207374_at | 0.0727 | 0.8078 | |
| GSE77861 | PLSCR2 | 57047 | 207374_at | 0.0714 | 0.6132 | |
| SRP133303 | PLSCR2 | 57047 | RNAseq | 0.4301 | 0.0834 | |
| SRP193095 | PLSCR2 | 57047 | RNAseq | 0.9517 | 0.0000 | |
| TCGA | PLSCR2 | 57047 | RNAseq | 0.7656 | 0.2915 |
Upregulated datasets: 0; Downregulated datasets: 0.
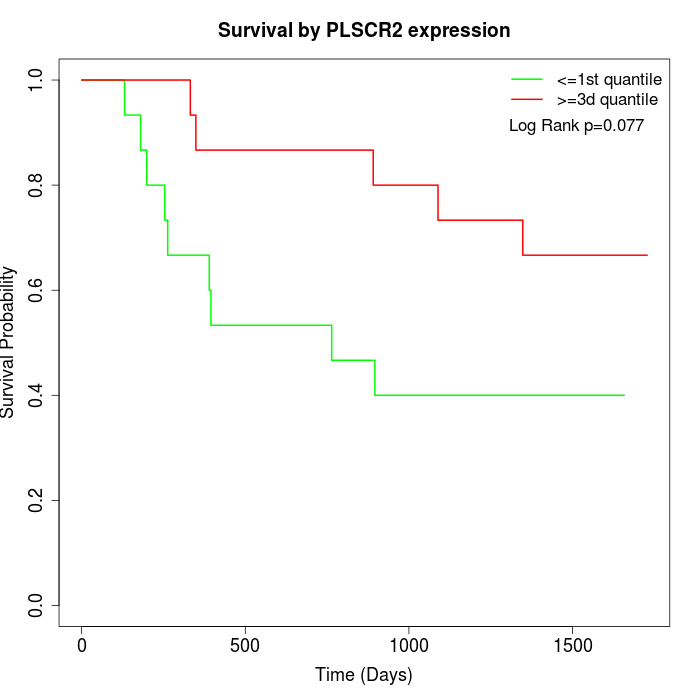
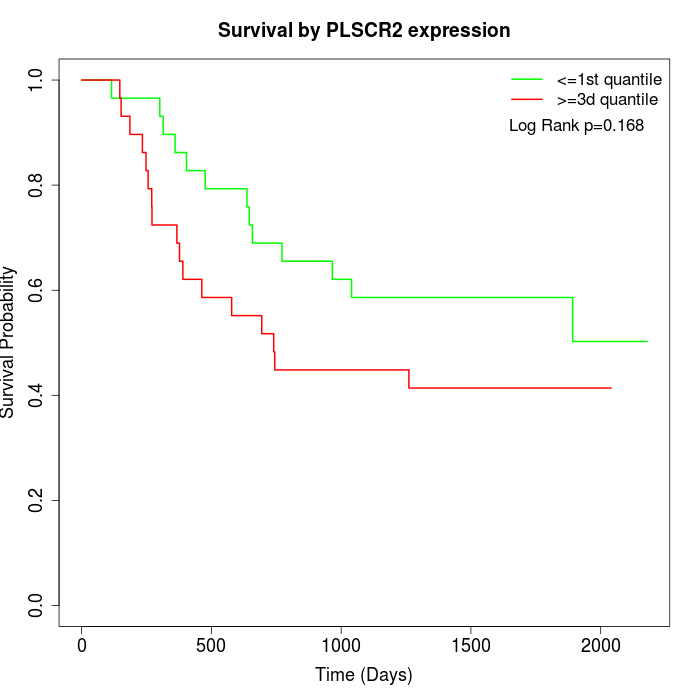
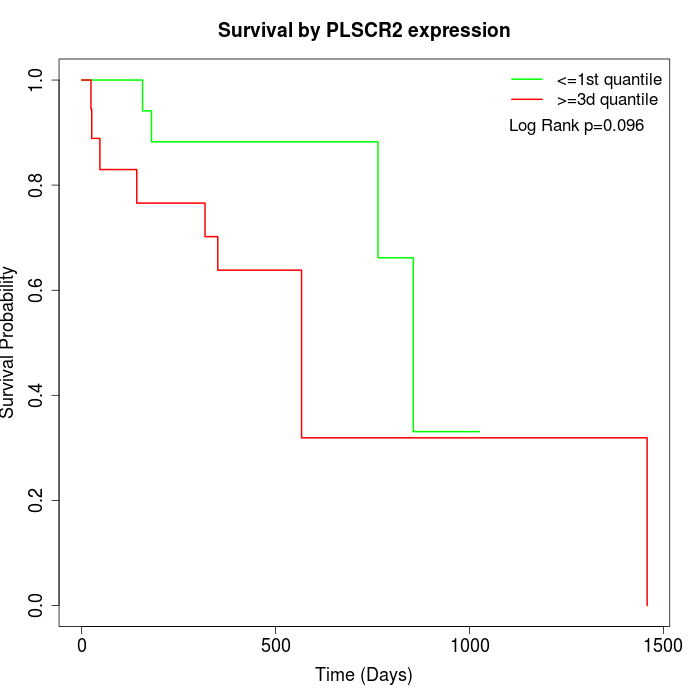
Survival by PLSCR2 expression:
Note: Click image to view full size file.
Copy number change of PLSCR2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PLSCR2 | 57047 | 22 | 0 | 8 | |
| GSE20123 | PLSCR2 | 57047 | 22 | 0 | 8 | |
| GSE43470 | PLSCR2 | 57047 | 23 | 0 | 20 | |
| GSE46452 | PLSCR2 | 57047 | 16 | 3 | 40 | |
| GSE47630 | PLSCR2 | 57047 | 19 | 3 | 18 | |
| GSE54993 | PLSCR2 | 57047 | 1 | 9 | 60 | |
| GSE54994 | PLSCR2 | 57047 | 34 | 1 | 18 | |
| GSE60625 | PLSCR2 | 57047 | 0 | 6 | 5 | |
| GSE74703 | PLSCR2 | 57047 | 20 | 0 | 16 | |
| GSE74704 | PLSCR2 | 57047 | 15 | 0 | 5 | |
| TCGA | PLSCR2 | 57047 | 67 | 2 | 27 |
Total number of gains: 239; Total number of losses: 24; Total Number of normals: 225.
Somatic mutations of PLSCR2:
Generating mutation plots.
Highly correlated genes for PLSCR2:
Showing top 20/109 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PLSCR2 | RFPL3 | 0.632948 | 5 | 0 | 5 |
| PLSCR2 | GNL3L | 0.63216 | 3 | 0 | 3 |
| PLSCR2 | CEP97 | 0.626004 | 5 | 0 | 3 |
| PLSCR2 | ITGA6 | 0.622307 | 3 | 0 | 3 |
| PLSCR2 | SLC30A3 | 0.614773 | 6 | 0 | 6 |
| PLSCR2 | EFCAB11 | 0.606431 | 3 | 0 | 3 |
| PLSCR2 | RCC2 | 0.60225 | 3 | 0 | 3 |
| PLSCR2 | KIR2DL2 | 0.596802 | 4 | 0 | 4 |
| PLSCR2 | BATF3 | 0.595608 | 3 | 0 | 3 |
| PLSCR2 | HDAC1 | 0.59422 | 3 | 0 | 3 |
| PLSCR2 | KMO | 0.592001 | 4 | 0 | 3 |
| PLSCR2 | EXTL3 | 0.587679 | 3 | 0 | 3 |
| PLSCR2 | PF4V1 | 0.579465 | 4 | 0 | 4 |
| PLSCR2 | CLCN2 | 0.576247 | 4 | 0 | 3 |
| PLSCR2 | XPNPEP3 | 0.576142 | 5 | 0 | 3 |
| PLSCR2 | CD80 | 0.575401 | 4 | 0 | 4 |
| PLSCR2 | LY96 | 0.573643 | 4 | 0 | 3 |
| PLSCR2 | GRK4 | 0.572204 | 4 | 0 | 4 |
| PLSCR2 | CCR8 | 0.571515 | 6 | 0 | 5 |
| PLSCR2 | OR10H1 | 0.571396 | 5 | 0 | 5 |
For details and further investigation, click here