| Full name: plexin B2 | Alias Symbol: MM1|KIAA0315|PLEXB2 | ||
| Type: protein-coding gene | Cytoband: 22q13.33 | ||
| Entrez ID: 23654 | HGNC ID: HGNC:9104 | Ensembl Gene: ENSG00000196576 | OMIM ID: 604293 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
PLXNB2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04360 | Axon guidance |
Expression of PLXNB2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PLXNB2 | 23654 | 208890_s_at | 0.0705 | 0.8664 | |
| GSE20347 | PLXNB2 | 23654 | 208890_s_at | -0.4274 | 0.0002 | |
| GSE23400 | PLXNB2 | 23654 | 208890_s_at | -0.0731 | 0.3148 | |
| GSE26886 | PLXNB2 | 23654 | 208890_s_at | 0.3237 | 0.2542 | |
| GSE29001 | PLXNB2 | 23654 | 208890_s_at | -0.4476 | 0.1550 | |
| GSE38129 | PLXNB2 | 23654 | 208890_s_at | -0.2638 | 0.0181 | |
| GSE45670 | PLXNB2 | 23654 | 208890_s_at | 0.0208 | 0.8971 | |
| GSE53622 | PLXNB2 | 23654 | 3723 | 0.0959 | 0.5623 | |
| GSE53624 | PLXNB2 | 23654 | 3723 | 0.0726 | 0.2839 | |
| GSE63941 | PLXNB2 | 23654 | 208890_s_at | -0.4450 | 0.5033 | |
| GSE77861 | PLXNB2 | 23654 | 208890_s_at | 0.1018 | 0.6907 | |
| GSE97050 | PLXNB2 | 23654 | A_33_P3321836 | 0.2155 | 0.2921 | |
| SRP007169 | PLXNB2 | 23654 | RNAseq | -0.4223 | 0.3381 | |
| SRP008496 | PLXNB2 | 23654 | RNAseq | -0.5193 | 0.0256 | |
| SRP064894 | PLXNB2 | 23654 | RNAseq | 0.2402 | 0.2527 | |
| SRP133303 | PLXNB2 | 23654 | RNAseq | 0.0449 | 0.7626 | |
| SRP159526 | PLXNB2 | 23654 | RNAseq | 0.1340 | 0.6700 | |
| SRP193095 | PLXNB2 | 23654 | RNAseq | -0.1297 | 0.4245 | |
| SRP219564 | PLXNB2 | 23654 | RNAseq | 0.0170 | 0.9665 | |
| TCGA | PLXNB2 | 23654 | RNAseq | -0.0924 | 0.0212 |
Upregulated datasets: 0; Downregulated datasets: 0.
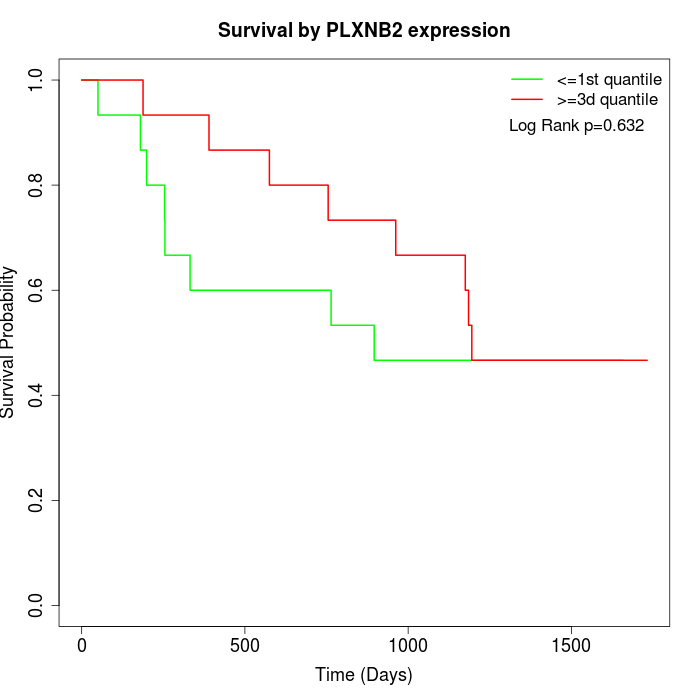
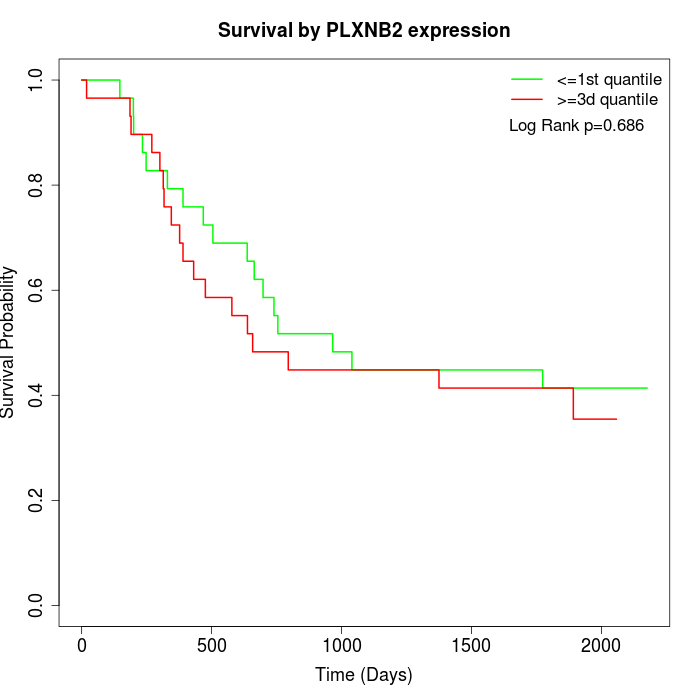
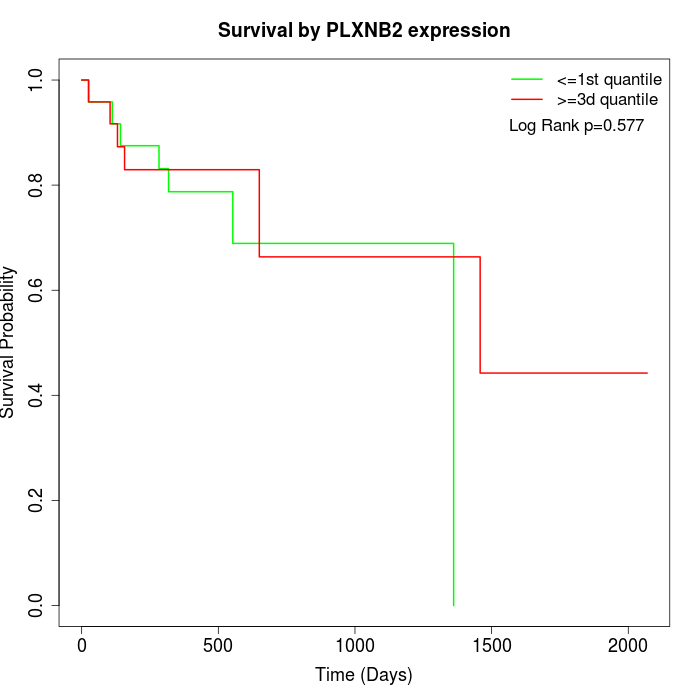
Survival by PLXNB2 expression:
Note: Click image to view full size file.
Copy number change of PLXNB2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PLXNB2 | 23654 | 3 | 8 | 19 | |
| GSE20123 | PLXNB2 | 23654 | 3 | 8 | 19 | |
| GSE43470 | PLXNB2 | 23654 | 2 | 8 | 33 | |
| GSE46452 | PLXNB2 | 23654 | 31 | 2 | 26 | |
| GSE47630 | PLXNB2 | 23654 | 9 | 4 | 27 | |
| GSE54993 | PLXNB2 | 23654 | 4 | 6 | 60 | |
| GSE54994 | PLXNB2 | 23654 | 11 | 8 | 34 | |
| GSE60625 | PLXNB2 | 23654 | 5 | 0 | 6 | |
| GSE74703 | PLXNB2 | 23654 | 2 | 6 | 28 | |
| GSE74704 | PLXNB2 | 23654 | 1 | 4 | 15 | |
| TCGA | PLXNB2 | 23654 | 26 | 16 | 54 |
Total number of gains: 97; Total number of losses: 70; Total Number of normals: 321.
Somatic mutations of PLXNB2:
Generating mutation plots.
Highly correlated genes for PLXNB2:
Showing top 20/238 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PLXNB2 | IFNGR2 | 0.753587 | 3 | 0 | 3 |
| PLXNB2 | NUDT14 | 0.748304 | 3 | 0 | 3 |
| PLXNB2 | SPG7 | 0.743283 | 3 | 0 | 3 |
| PLXNB2 | MKNK1 | 0.728188 | 3 | 0 | 3 |
| PLXNB2 | RNF187 | 0.727355 | 4 | 0 | 4 |
| PLXNB2 | RBCK1 | 0.725903 | 3 | 0 | 3 |
| PLXNB2 | RAC3 | 0.711765 | 3 | 0 | 3 |
| PLXNB2 | ZNF408 | 0.709733 | 3 | 0 | 3 |
| PLXNB2 | ATP13A1 | 0.707023 | 3 | 0 | 3 |
| PLXNB2 | TMEM54 | 0.705782 | 4 | 0 | 3 |
| PLXNB2 | ZGPAT | 0.701627 | 3 | 0 | 3 |
| PLXNB2 | VPS28 | 0.701415 | 3 | 0 | 3 |
| PLXNB2 | HNRNPUL1 | 0.701379 | 3 | 0 | 3 |
| PLXNB2 | UCKL1 | 0.694508 | 4 | 0 | 3 |
| PLXNB2 | DPYD | 0.694502 | 3 | 0 | 3 |
| PLXNB2 | SHANK3 | 0.693121 | 3 | 0 | 3 |
| PLXNB2 | NAGLU | 0.691364 | 3 | 0 | 3 |
| PLXNB2 | ELFN1 | 0.683242 | 3 | 0 | 3 |
| PLXNB2 | ARHGAP17 | 0.679616 | 4 | 0 | 4 |
| PLXNB2 | WTIP | 0.67655 | 3 | 0 | 3 |
For details and further investigation, click here