| Full name: PMS1 homolog 1, mismatch repair system component | Alias Symbol: MLH2 | ||
| Type: protein-coding gene | Cytoband: 2q32.2 | ||
| Entrez ID: 5378 | HGNC ID: HGNC:9121 | Ensembl Gene: ENSG00000064933 | OMIM ID: 600258 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of PMS1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PMS1 | 5378 | 213677_s_at | 0.4858 | 0.1778 | |
| GSE20347 | PMS1 | 5378 | 213677_s_at | 0.5494 | 0.0055 | |
| GSE23400 | PMS1 | 5378 | 213677_s_at | 0.4510 | 0.0001 | |
| GSE26886 | PMS1 | 5378 | 213677_s_at | 0.3741 | 0.2300 | |
| GSE29001 | PMS1 | 5378 | 213677_s_at | 0.4652 | 0.2231 | |
| GSE38129 | PMS1 | 5378 | 213677_s_at | 0.4996 | 0.0016 | |
| GSE45670 | PMS1 | 5378 | 213677_s_at | 0.1008 | 0.6031 | |
| GSE53622 | PMS1 | 5378 | 102948 | 0.1005 | 0.3365 | |
| GSE53624 | PMS1 | 5378 | 34968 | 0.3181 | 0.0000 | |
| GSE63941 | PMS1 | 5378 | 213677_s_at | 0.1830 | 0.7976 | |
| GSE77861 | PMS1 | 5378 | 213677_s_at | 0.2896 | 0.4764 | |
| GSE97050 | PMS1 | 5378 | A_33_P3250463 | 0.1122 | 0.7684 | |
| SRP007169 | PMS1 | 5378 | RNAseq | 0.4313 | 0.3513 | |
| SRP008496 | PMS1 | 5378 | RNAseq | 0.3262 | 0.2265 | |
| SRP064894 | PMS1 | 5378 | RNAseq | 0.3177 | 0.0748 | |
| SRP133303 | PMS1 | 5378 | RNAseq | 0.0542 | 0.6342 | |
| SRP159526 | PMS1 | 5378 | RNAseq | -0.2192 | 0.3603 | |
| SRP193095 | PMS1 | 5378 | RNAseq | -0.1091 | 0.2063 | |
| SRP219564 | PMS1 | 5378 | RNAseq | -0.0711 | 0.8109 | |
| TCGA | PMS1 | 5378 | RNAseq | 0.0375 | 0.5338 |
Upregulated datasets: 0; Downregulated datasets: 0.
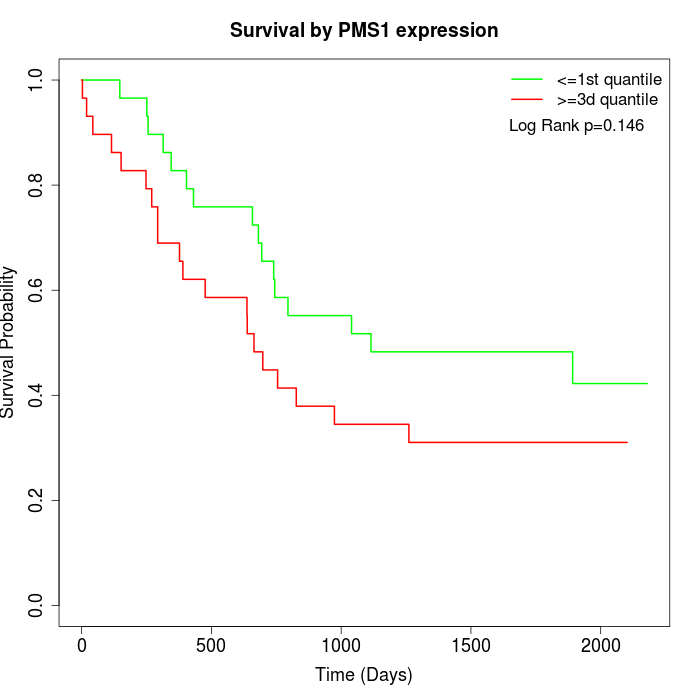
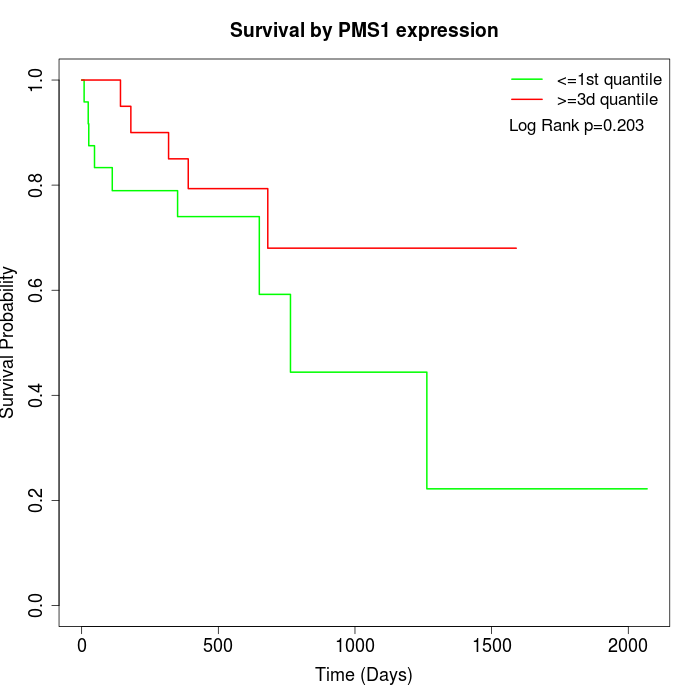
Survival by PMS1 expression:
Note: Click image to view full size file.
Copy number change of PMS1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PMS1 | 5378 | 9 | 3 | 18 | |
| GSE20123 | PMS1 | 5378 | 9 | 3 | 18 | |
| GSE43470 | PMS1 | 5378 | 3 | 1 | 39 | |
| GSE46452 | PMS1 | 5378 | 1 | 4 | 54 | |
| GSE47630 | PMS1 | 5378 | 4 | 5 | 31 | |
| GSE54993 | PMS1 | 5378 | 0 | 5 | 65 | |
| GSE54994 | PMS1 | 5378 | 12 | 6 | 35 | |
| GSE60625 | PMS1 | 5378 | 0 | 3 | 8 | |
| GSE74703 | PMS1 | 5378 | 2 | 1 | 33 | |
| GSE74704 | PMS1 | 5378 | 3 | 3 | 14 | |
| TCGA | PMS1 | 5378 | 24 | 7 | 65 |
Total number of gains: 67; Total number of losses: 41; Total Number of normals: 380.
Somatic mutations of PMS1:
Generating mutation plots.
Highly correlated genes for PMS1:
Showing top 20/701 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PMS1 | CISD2 | 0.815036 | 3 | 0 | 3 |
| PMS1 | CMTM4 | 0.814316 | 3 | 0 | 3 |
| PMS1 | TMEM181 | 0.811606 | 3 | 0 | 3 |
| PMS1 | DHRSX | 0.796291 | 3 | 0 | 3 |
| PMS1 | CHD8 | 0.79182 | 3 | 0 | 3 |
| PMS1 | CCDC127 | 0.780707 | 3 | 0 | 3 |
| PMS1 | SLC25A37 | 0.757476 | 3 | 0 | 3 |
| PMS1 | EVL | 0.751386 | 3 | 0 | 3 |
| PMS1 | CLPTM1L | 0.744639 | 3 | 0 | 3 |
| PMS1 | GAA | 0.743623 | 3 | 0 | 3 |
| PMS1 | RNF217 | 0.742946 | 3 | 0 | 3 |
| PMS1 | SFXN4 | 0.739255 | 3 | 0 | 3 |
| PMS1 | LARP7 | 0.737108 | 3 | 0 | 3 |
| PMS1 | SLC7A6 | 0.734785 | 3 | 0 | 3 |
| PMS1 | HPS5 | 0.722753 | 3 | 0 | 3 |
| PMS1 | GAL | 0.719244 | 3 | 0 | 3 |
| PMS1 | PDHX | 0.717441 | 3 | 0 | 3 |
| PMS1 | CHIC1 | 0.716857 | 3 | 0 | 3 |
| PMS1 | IQCE | 0.714315 | 3 | 0 | 3 |
| PMS1 | CELF1 | 0.710039 | 5 | 0 | 5 |
For details and further investigation, click here