| Full name: pyruvate dehydrogenase complex component X | Alias Symbol: E3BP|proX|PDX1|OPDX|DLDBP | ||
| Type: protein-coding gene | Cytoband: 11p13 | ||
| Entrez ID: 8050 | HGNC ID: HGNC:21350 | Ensembl Gene: ENSG00000110435 | OMIM ID: 608769 |
| Drug and gene relationship at DGIdb | |||
Expression of PDHX:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PDHX | 8050 | 203067_at | 0.1237 | 0.7927 | |
| GSE20347 | PDHX | 8050 | 203067_at | 0.0652 | 0.7330 | |
| GSE23400 | PDHX | 8050 | 203067_at | 0.2972 | 0.0005 | |
| GSE26886 | PDHX | 8050 | 203067_at | -0.0173 | 0.9617 | |
| GSE29001 | PDHX | 8050 | 203067_at | 0.5220 | 0.0755 | |
| GSE38129 | PDHX | 8050 | 203067_at | 0.0010 | 0.9955 | |
| GSE45670 | PDHX | 8050 | 203067_at | 0.1226 | 0.4315 | |
| GSE53622 | PDHX | 8050 | 55638 | -0.1011 | 0.1769 | |
| GSE53624 | PDHX | 8050 | 55638 | 0.0108 | 0.8929 | |
| GSE63941 | PDHX | 8050 | 203067_at | -0.7177 | 0.1400 | |
| GSE77861 | PDHX | 8050 | 203067_at | 0.2957 | 0.4290 | |
| GSE97050 | PDHX | 8050 | A_23_P36266 | -0.1908 | 0.4343 | |
| SRP007169 | PDHX | 8050 | RNAseq | 0.0666 | 0.8727 | |
| SRP008496 | PDHX | 8050 | RNAseq | 0.0988 | 0.7104 | |
| SRP064894 | PDHX | 8050 | RNAseq | -0.0205 | 0.9010 | |
| SRP133303 | PDHX | 8050 | RNAseq | 0.3327 | 0.0299 | |
| SRP159526 | PDHX | 8050 | RNAseq | 0.1553 | 0.5102 | |
| SRP193095 | PDHX | 8050 | RNAseq | -0.1057 | 0.3161 | |
| SRP219564 | PDHX | 8050 | RNAseq | 0.1068 | 0.7518 | |
| TCGA | PDHX | 8050 | RNAseq | -0.1709 | 0.0041 |
Upregulated datasets: 0; Downregulated datasets: 0.
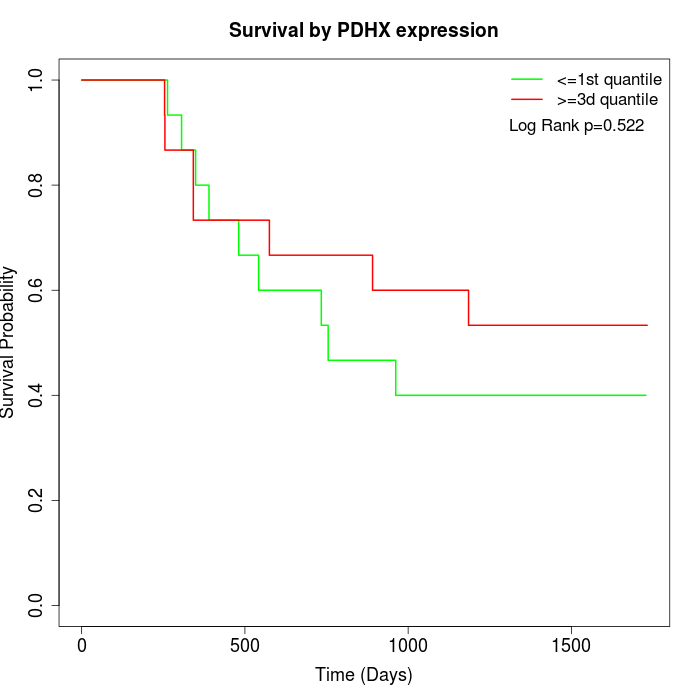
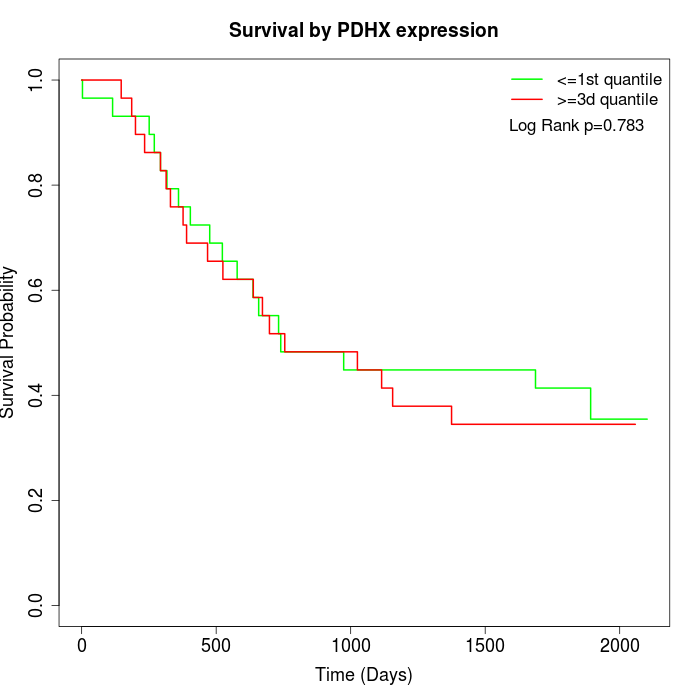
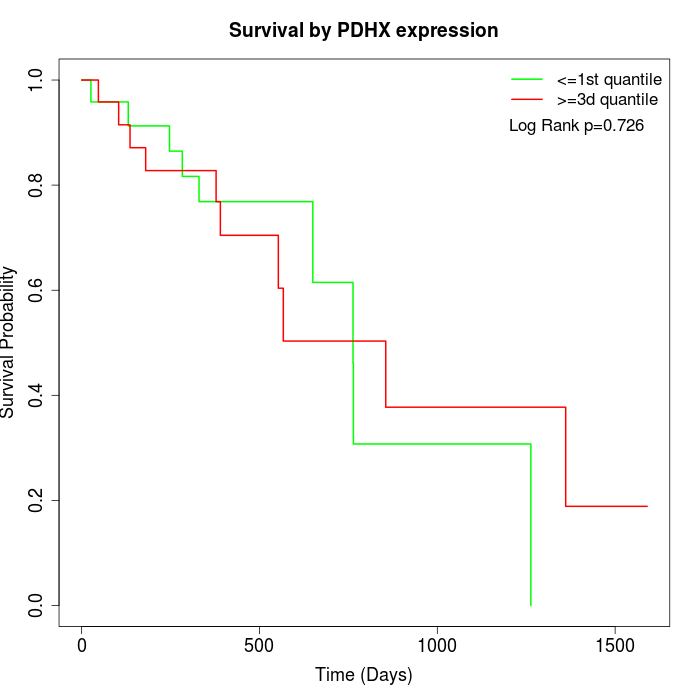
Survival by PDHX expression:
Note: Click image to view full size file.
Copy number change of PDHX:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PDHX | 8050 | 1 | 8 | 21 | |
| GSE20123 | PDHX | 8050 | 1 | 7 | 22 | |
| GSE43470 | PDHX | 8050 | 3 | 4 | 36 | |
| GSE46452 | PDHX | 8050 | 7 | 5 | 47 | |
| GSE47630 | PDHX | 8050 | 3 | 10 | 27 | |
| GSE54993 | PDHX | 8050 | 3 | 2 | 65 | |
| GSE54994 | PDHX | 8050 | 3 | 10 | 40 | |
| GSE60625 | PDHX | 8050 | 0 | 0 | 11 | |
| GSE74703 | PDHX | 8050 | 2 | 2 | 32 | |
| GSE74704 | PDHX | 8050 | 0 | 4 | 16 | |
| TCGA | PDHX | 8050 | 16 | 23 | 57 |
Total number of gains: 39; Total number of losses: 75; Total Number of normals: 374.
Somatic mutations of PDHX:
Generating mutation plots.
Highly correlated genes for PDHX:
Showing top 20/458 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PDHX | RBMS1 | 0.816227 | 3 | 0 | 3 |
| PDHX | ANAPC4 | 0.812691 | 3 | 0 | 3 |
| PDHX | KPNA5 | 0.792327 | 3 | 0 | 3 |
| PDHX | C9orf64 | 0.762934 | 3 | 0 | 3 |
| PDHX | REEP5 | 0.748278 | 3 | 0 | 3 |
| PDHX | SLC22A5 | 0.747246 | 3 | 0 | 3 |
| PDHX | KIAA1143 | 0.739322 | 3 | 0 | 3 |
| PDHX | AP1AR | 0.734575 | 3 | 0 | 3 |
| PDHX | LARS2 | 0.73397 | 3 | 0 | 3 |
| PDHX | MED7 | 0.727703 | 3 | 0 | 3 |
| PDHX | CWC27 | 0.725871 | 3 | 0 | 3 |
| PDHX | TM9SF1 | 0.725775 | 3 | 0 | 3 |
| PDHX | C15orf61 | 0.725553 | 3 | 0 | 3 |
| PDHX | PMS1 | 0.717441 | 3 | 0 | 3 |
| PDHX | CETN3 | 0.714258 | 3 | 0 | 3 |
| PDHX | ZZZ3 | 0.707355 | 4 | 0 | 3 |
| PDHX | PON2 | 0.701936 | 3 | 0 | 3 |
| PDHX | CBLL1 | 0.70006 | 4 | 0 | 3 |
| PDHX | ANGEL2 | 0.698358 | 3 | 0 | 3 |
| PDHX | DYRK2 | 0.695524 | 3 | 0 | 3 |
For details and further investigation, click here