| Full name: protein phosphatase, Mg2+/Mn2+ dependent 1N (putative) | Alias Symbol: FLJ40125 | ||
| Type: protein-coding gene | Cytoband: 19q13.32 | ||
| Entrez ID: 147699 | HGNC ID: HGNC:26845 | Ensembl Gene: ENSG00000213889 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of PPM1N:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PPM1N | 147699 | 229559_at | -0.2078 | 0.4630 | |
| GSE26886 | PPM1N | 147699 | 229559_at | 0.0583 | 0.7238 | |
| GSE45670 | PPM1N | 147699 | 229559_at | 0.0783 | 0.7110 | |
| GSE53622 | PPM1N | 147699 | 41318 | 0.2726 | 0.0982 | |
| GSE53624 | PPM1N | 147699 | 41318 | 0.4158 | 0.0000 | |
| GSE63941 | PPM1N | 147699 | 229559_at | -0.4913 | 0.3043 | |
| GSE77861 | PPM1N | 147699 | 229559_at | -0.1622 | 0.1097 | |
| GSE97050 | PPM1N | 147699 | A_33_P3279681 | 0.0449 | 0.8315 | |
| SRP064894 | PPM1N | 147699 | RNAseq | 0.0206 | 0.9501 | |
| SRP133303 | PPM1N | 147699 | RNAseq | 0.0627 | 0.7672 | |
| SRP159526 | PPM1N | 147699 | RNAseq | -0.2260 | 0.6079 | |
| SRP193095 | PPM1N | 147699 | RNAseq | -0.0151 | 0.9339 | |
| SRP219564 | PPM1N | 147699 | RNAseq | -0.4046 | 0.3723 | |
| TCGA | PPM1N | 147699 | RNAseq | 0.2475 | 0.3280 |
Upregulated datasets: 0; Downregulated datasets: 0.
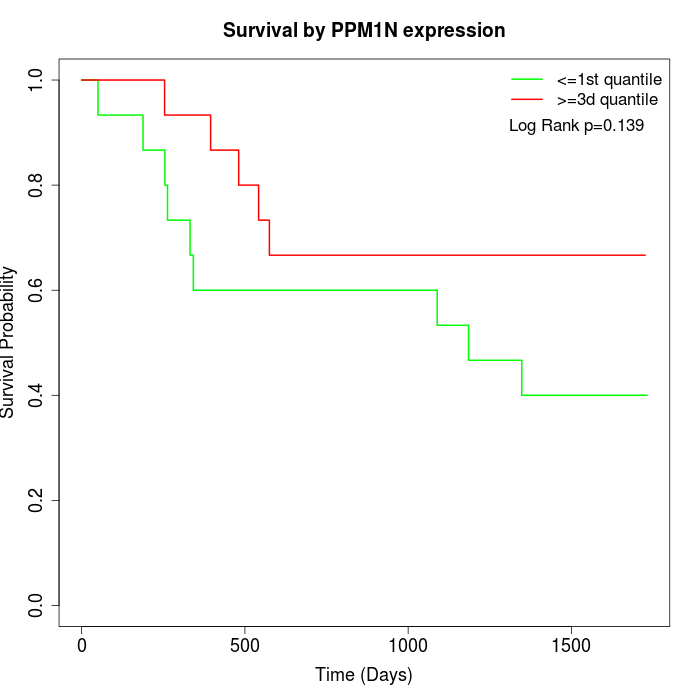
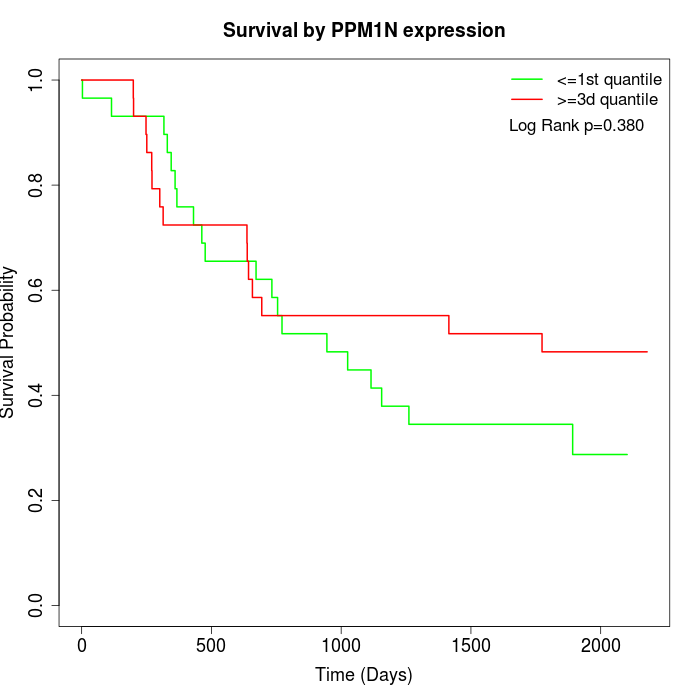
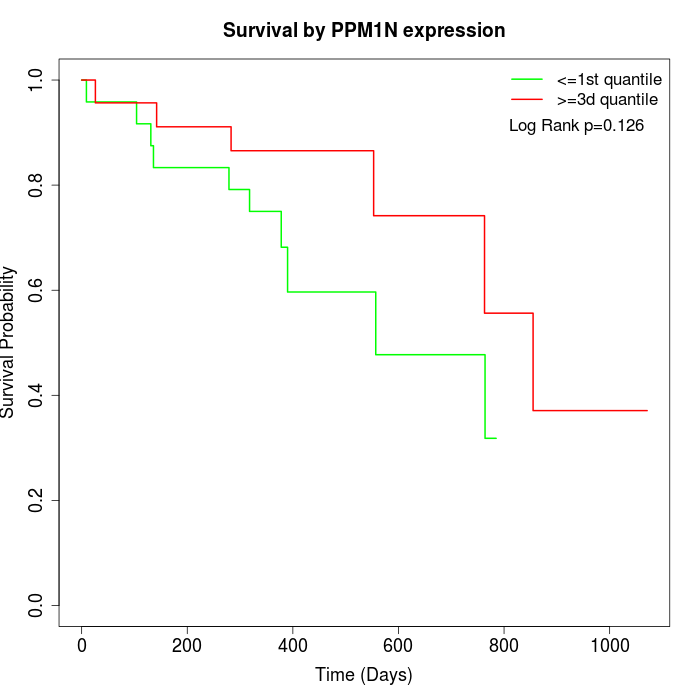
Survival by PPM1N expression:
Note: Click image to view full size file.
Copy number change of PPM1N:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PPM1N | 147699 | 4 | 5 | 21 | |
| GSE20123 | PPM1N | 147699 | 4 | 4 | 22 | |
| GSE43470 | PPM1N | 147699 | 3 | 11 | 29 | |
| GSE46452 | PPM1N | 147699 | 45 | 1 | 13 | |
| GSE47630 | PPM1N | 147699 | 8 | 6 | 26 | |
| GSE54993 | PPM1N | 147699 | 17 | 4 | 49 | |
| GSE54994 | PPM1N | 147699 | 6 | 12 | 35 | |
| GSE60625 | PPM1N | 147699 | 9 | 0 | 2 | |
| GSE74703 | PPM1N | 147699 | 3 | 7 | 26 | |
| GSE74704 | PPM1N | 147699 | 4 | 2 | 14 | |
| TCGA | PPM1N | 147699 | 15 | 16 | 65 |
Total number of gains: 118; Total number of losses: 68; Total Number of normals: 302.
Somatic mutations of PPM1N:
Generating mutation plots.
Highly correlated genes for PPM1N:
Showing top 20/45 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PPM1N | KCNMB2 | 0.746439 | 3 | 0 | 3 |
| PPM1N | AVPR1B | 0.744505 | 3 | 0 | 3 |
| PPM1N | C11orf21 | 0.733946 | 3 | 0 | 3 |
| PPM1N | THRSP | 0.733316 | 3 | 0 | 3 |
| PPM1N | A1CF | 0.70055 | 3 | 0 | 3 |
| PPM1N | ADAMTS13 | 0.698472 | 3 | 0 | 3 |
| PPM1N | SPRY3 | 0.697668 | 3 | 0 | 3 |
| PPM1N | SLC22A14 | 0.69058 | 3 | 0 | 3 |
| PPM1N | CCDC33 | 0.68895 | 3 | 0 | 3 |
| PPM1N | PNPLA7 | 0.678739 | 3 | 0 | 3 |
| PPM1N | OR1J4 | 0.678329 | 3 | 0 | 3 |
| PPM1N | EML2 | 0.673782 | 3 | 0 | 3 |
| PPM1N | PIWIL2 | 0.672532 | 3 | 0 | 3 |
| PPM1N | TNXB | 0.655675 | 3 | 0 | 3 |
| PPM1N | MUC8 | 0.648493 | 3 | 0 | 3 |
| PPM1N | FOXB1 | 0.64802 | 3 | 0 | 3 |
| PPM1N | SPTBN4 | 0.647858 | 4 | 0 | 3 |
| PPM1N | AMN | 0.644076 | 3 | 0 | 3 |
| PPM1N | NOXA1 | 0.641357 | 3 | 0 | 3 |
| PPM1N | CDHR2 | 0.640795 | 4 | 0 | 3 |
For details and further investigation, click here