| Full name: DEAH-box helicase 35 | Alias Symbol: FLJ22759|KAIA0875 | ||
| Type: protein-coding gene | Cytoband: 20q11.23-q12 | ||
| Entrez ID: 60625 | HGNC ID: HGNC:15861 | Ensembl Gene: ENSG00000101452 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of DHX35:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DHX35 | 60625 | 218579_s_at | 0.5363 | 0.2652 | |
| GSE20347 | DHX35 | 60625 | 218579_s_at | 0.5887 | 0.0020 | |
| GSE23400 | DHX35 | 60625 | 218579_s_at | 0.1121 | 0.2210 | |
| GSE26886 | DHX35 | 60625 | 218579_s_at | -1.0993 | 0.0016 | |
| GSE29001 | DHX35 | 60625 | 218579_s_at | 0.3143 | 0.2219 | |
| GSE38129 | DHX35 | 60625 | 218579_s_at | 0.5060 | 0.0035 | |
| GSE45670 | DHX35 | 60625 | 218579_s_at | 0.1934 | 0.5097 | |
| GSE53622 | DHX35 | 60625 | 4050 | 0.3456 | 0.0000 | |
| GSE53624 | DHX35 | 60625 | 4050 | 0.2908 | 0.0000 | |
| GSE63941 | DHX35 | 60625 | 234728_s_at | 3.2166 | 0.0000 | |
| GSE77861 | DHX35 | 60625 | 218579_s_at | -0.0461 | 0.9146 | |
| GSE97050 | DHX35 | 60625 | A_23_P5945 | 0.1430 | 0.4880 | |
| SRP007169 | DHX35 | 60625 | RNAseq | 1.0646 | 0.0616 | |
| SRP008496 | DHX35 | 60625 | RNAseq | 0.2212 | 0.6099 | |
| SRP064894 | DHX35 | 60625 | RNAseq | 0.0432 | 0.8135 | |
| SRP133303 | DHX35 | 60625 | RNAseq | 0.5224 | 0.0199 | |
| SRP159526 | DHX35 | 60625 | RNAseq | 0.0429 | 0.8961 | |
| SRP193095 | DHX35 | 60625 | RNAseq | -0.5679 | 0.0001 | |
| SRP219564 | DHX35 | 60625 | RNAseq | 0.3173 | 0.3901 | |
| TCGA | DHX35 | 60625 | RNAseq | 0.0886 | 0.1498 |
Upregulated datasets: 1; Downregulated datasets: 1.
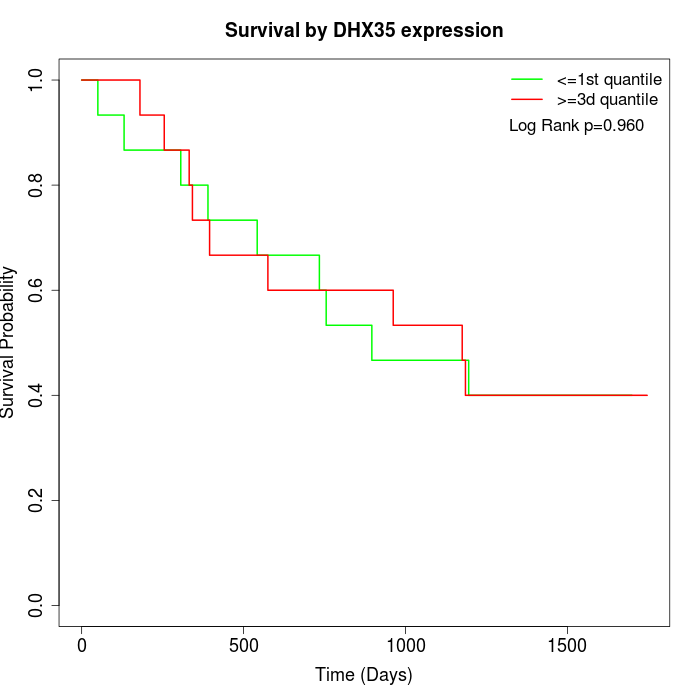
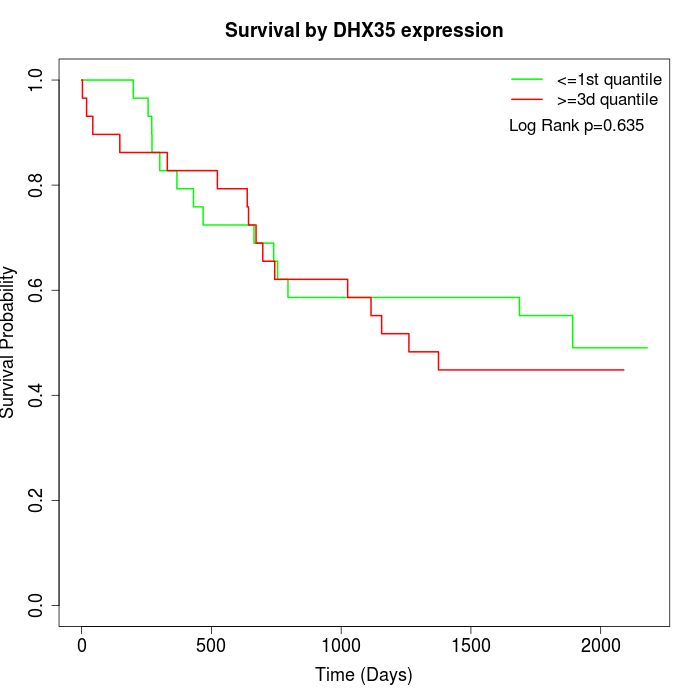
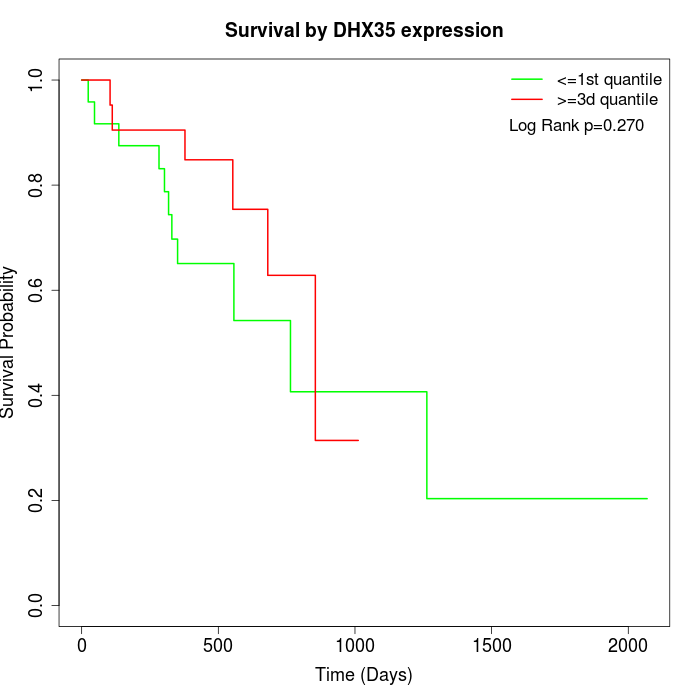
Survival by DHX35 expression:
Note: Click image to view full size file.
Copy number change of DHX35:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DHX35 | 60625 | 14 | 2 | 14 | |
| GSE20123 | DHX35 | 60625 | 14 | 2 | 14 | |
| GSE43470 | DHX35 | 60625 | 13 | 0 | 30 | |
| GSE46452 | DHX35 | 60625 | 29 | 0 | 30 | |
| GSE47630 | DHX35 | 60625 | 24 | 0 | 16 | |
| GSE54993 | DHX35 | 60625 | 0 | 17 | 53 | |
| GSE54994 | DHX35 | 60625 | 24 | 0 | 29 | |
| GSE60625 | DHX35 | 60625 | 0 | 0 | 11 | |
| GSE74703 | DHX35 | 60625 | 10 | 0 | 26 | |
| GSE74704 | DHX35 | 60625 | 10 | 1 | 9 | |
| TCGA | DHX35 | 60625 | 47 | 4 | 45 |
Total number of gains: 185; Total number of losses: 26; Total Number of normals: 277.
Somatic mutations of DHX35:
Generating mutation plots.
Highly correlated genes for DHX35:
Showing top 20/270 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DHX35 | ACACA | 0.781387 | 3 | 0 | 3 |
| DHX35 | PPTC7 | 0.780876 | 3 | 0 | 3 |
| DHX35 | PRKAG1 | 0.778084 | 3 | 0 | 3 |
| DHX35 | TYSND1 | 0.765367 | 3 | 0 | 3 |
| DHX35 | NXF1 | 0.739901 | 3 | 0 | 3 |
| DHX35 | SRP68 | 0.732701 | 3 | 0 | 3 |
| DHX35 | CCDC85C | 0.73098 | 3 | 0 | 3 |
| DHX35 | TP53RK | 0.728797 | 3 | 0 | 3 |
| DHX35 | CYCS | 0.727462 | 3 | 0 | 3 |
| DHX35 | C1orf115 | 0.727208 | 3 | 0 | 3 |
| DHX35 | PRKCH | 0.717683 | 3 | 0 | 3 |
| DHX35 | TXLNG | 0.717263 | 3 | 0 | 3 |
| DHX35 | GABPA | 0.713493 | 3 | 0 | 3 |
| DHX35 | CHD7 | 0.708817 | 3 | 0 | 3 |
| DHX35 | SUPT4H1 | 0.708322 | 3 | 0 | 3 |
| DHX35 | ZCCHC8 | 0.702181 | 3 | 0 | 3 |
| DHX35 | PNP | 0.699528 | 3 | 0 | 3 |
| DHX35 | SLC25A39 | 0.696506 | 3 | 0 | 3 |
| DHX35 | KLF13 | 0.691072 | 3 | 0 | 3 |
| DHX35 | S1PR5 | 0.690186 | 3 | 0 | 3 |
For details and further investigation, click here