| Full name: protein kinase AMP-activated non-catalytic subunit gamma 2 | Alias Symbol: AAKG|AAKG2|H91620p|WPWS|CMH6 | ||
| Type: protein-coding gene | Cytoband: 7q36.1 | ||
| Entrez ID: 51422 | HGNC ID: HGNC:9386 | Ensembl Gene: ENSG00000106617 | OMIM ID: 602743 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
PRKAG2 involved pathways:
Expression of PRKAG2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PRKAG2 | 51422 | 233748_x_at | -0.6679 | 0.3115 | |
| GSE20347 | PRKAG2 | 51422 | 218292_s_at | 0.0654 | 0.5484 | |
| GSE23400 | PRKAG2 | 51422 | 218292_s_at | -0.0436 | 0.3850 | |
| GSE26886 | PRKAG2 | 51422 | 231224_x_at | 0.0999 | 0.2818 | |
| GSE29001 | PRKAG2 | 51422 | 218292_s_at | -0.3549 | 0.0138 | |
| GSE38129 | PRKAG2 | 51422 | 218292_s_at | -0.1670 | 0.2794 | |
| GSE45670 | PRKAG2 | 51422 | 222582_at | -0.7031 | 0.0052 | |
| GSE53622 | PRKAG2 | 51422 | 74146 | -0.4846 | 0.0000 | |
| GSE53624 | PRKAG2 | 51422 | 63835 | -0.2101 | 0.0031 | |
| GSE63941 | PRKAG2 | 51422 | 233748_x_at | -1.0238 | 0.0229 | |
| GSE77861 | PRKAG2 | 51422 | 222582_at | 0.1725 | 0.2931 | |
| GSE97050 | PRKAG2 | 51422 | A_23_P314760 | -0.2010 | 0.6431 | |
| SRP007169 | PRKAG2 | 51422 | RNAseq | 1.3768 | 0.0074 | |
| SRP008496 | PRKAG2 | 51422 | RNAseq | 0.7644 | 0.0233 | |
| SRP064894 | PRKAG2 | 51422 | RNAseq | -0.4111 | 0.0450 | |
| SRP133303 | PRKAG2 | 51422 | RNAseq | -0.1029 | 0.5947 | |
| SRP159526 | PRKAG2 | 51422 | RNAseq | -0.2001 | 0.5698 | |
| SRP193095 | PRKAG2 | 51422 | RNAseq | 0.0472 | 0.7942 | |
| SRP219564 | PRKAG2 | 51422 | RNAseq | 0.0663 | 0.9133 | |
| TCGA | PRKAG2 | 51422 | RNAseq | -0.4316 | 0.0000 |
Upregulated datasets: 1; Downregulated datasets: 1.
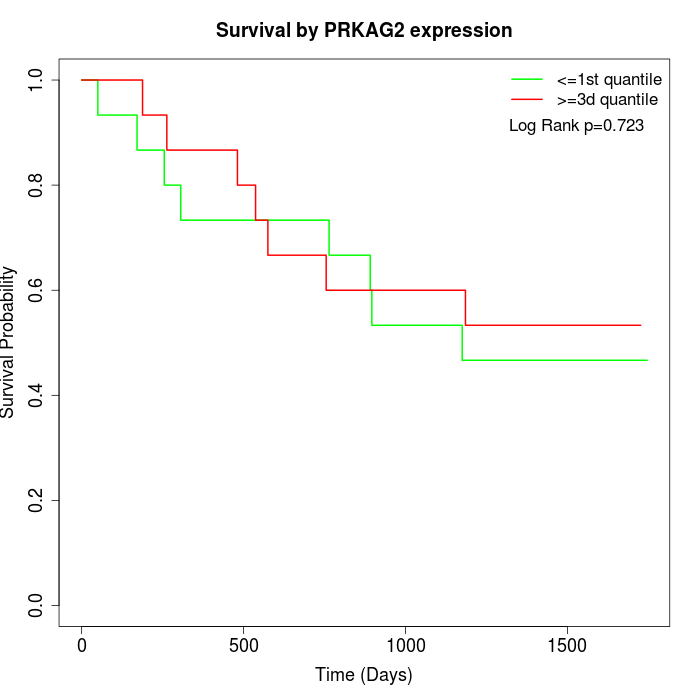
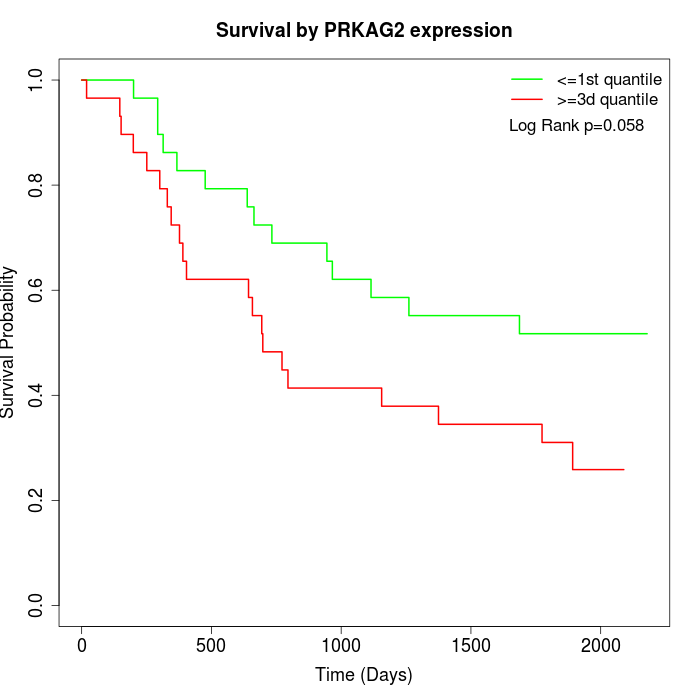
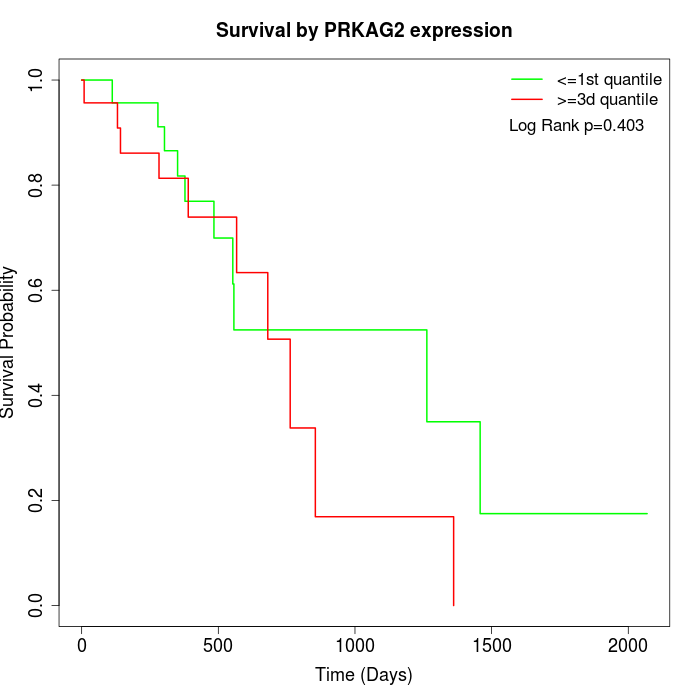
Survival by PRKAG2 expression:
Note: Click image to view full size file.
Copy number change of PRKAG2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PRKAG2 | 51422 | 2 | 4 | 24 | |
| GSE20123 | PRKAG2 | 51422 | 2 | 4 | 24 | |
| GSE43470 | PRKAG2 | 51422 | 2 | 5 | 36 | |
| GSE46452 | PRKAG2 | 51422 | 7 | 2 | 50 | |
| GSE47630 | PRKAG2 | 51422 | 6 | 9 | 25 | |
| GSE54993 | PRKAG2 | 51422 | 3 | 3 | 64 | |
| GSE54994 | PRKAG2 | 51422 | 5 | 8 | 40 | |
| GSE60625 | PRKAG2 | 51422 | 0 | 0 | 11 | |
| GSE74703 | PRKAG2 | 51422 | 2 | 4 | 30 | |
| GSE74704 | PRKAG2 | 51422 | 1 | 4 | 15 | |
| TCGA | PRKAG2 | 51422 | 27 | 26 | 43 |
Total number of gains: 57; Total number of losses: 69; Total Number of normals: 362.
Somatic mutations of PRKAG2:
Generating mutation plots.
Highly correlated genes for PRKAG2:
Showing top 20/632 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PRKAG2 | EID3 | 0.805635 | 3 | 0 | 3 |
| PRKAG2 | ADCY6 | 0.748746 | 3 | 0 | 3 |
| PRKAG2 | PNKD | 0.726293 | 3 | 0 | 3 |
| PRKAG2 | CCDC136 | 0.723518 | 3 | 0 | 3 |
| PRKAG2 | MYCT1 | 0.722992 | 3 | 0 | 3 |
| PRKAG2 | SYNC | 0.722402 | 5 | 0 | 5 |
| PRKAG2 | ING4 | 0.718417 | 3 | 0 | 3 |
| PRKAG2 | UBXN10 | 0.717913 | 3 | 0 | 3 |
| PRKAG2 | ZBTB26 | 0.715673 | 3 | 0 | 3 |
| PRKAG2 | IGSF10 | 0.713635 | 3 | 0 | 3 |
| PRKAG2 | RHEB | 0.707772 | 6 | 0 | 6 |
| PRKAG2 | RNF130 | 0.701427 | 3 | 0 | 3 |
| PRKAG2 | CCDC121 | 0.700364 | 3 | 0 | 3 |
| PRKAG2 | FIBIN | 0.700001 | 4 | 0 | 4 |
| PRKAG2 | CHIC1 | 0.699983 | 3 | 0 | 3 |
| PRKAG2 | N6AMT1 | 0.696572 | 4 | 0 | 4 |
| PRKAG2 | DNALI1 | 0.694735 | 6 | 0 | 5 |
| PRKAG2 | ECSCR | 0.694544 | 3 | 0 | 3 |
| PRKAG2 | PODN | 0.693849 | 4 | 0 | 3 |
| PRKAG2 | NEK7 | 0.69335 | 3 | 0 | 3 |
For details and further investigation, click here