| Full name: protein kinase X-linked | Alias Symbol: PKX1 | ||
| Type: protein-coding gene | Cytoband: Xp22.33 | ||
| Entrez ID: 5613 | HGNC ID: HGNC:9441 | Ensembl Gene: ENSG00000183943 | OMIM ID: 300083 |
| Drug and gene relationship at DGIdb | |||
Expression of PRKX:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PRKX | 5613 | 204061_at | 0.4512 | 0.6051 | |
| GSE20347 | PRKX | 5613 | 204061_at | -0.2456 | 0.5548 | |
| GSE23400 | PRKX | 5613 | 204061_at | 0.2156 | 0.1429 | |
| GSE26886 | PRKX | 5613 | 204061_at | -1.1449 | 0.0260 | |
| GSE29001 | PRKX | 5613 | 204061_at | -0.2359 | 0.7291 | |
| GSE38129 | PRKX | 5613 | 204061_at | 0.3109 | 0.4205 | |
| GSE45670 | PRKX | 5613 | 204061_at | -0.0849 | 0.8593 | |
| GSE53622 | PRKX | 5613 | 59533 | -0.1580 | 0.3851 | |
| GSE53624 | PRKX | 5613 | 59533 | -0.0786 | 0.5721 | |
| GSE63941 | PRKX | 5613 | 204061_at | 0.0257 | 0.9798 | |
| GSE77861 | PRKX | 5613 | 204061_at | 0.8140 | 0.0177 | |
| GSE97050 | PRKX | 5613 | A_23_P217339 | -0.1693 | 0.5726 | |
| SRP007169 | PRKX | 5613 | RNAseq | 0.6637 | 0.2178 | |
| SRP008496 | PRKX | 5613 | RNAseq | 0.5201 | 0.1012 | |
| SRP064894 | PRKX | 5613 | RNAseq | -0.5041 | 0.0062 | |
| SRP133303 | PRKX | 5613 | RNAseq | 0.0378 | 0.9107 | |
| SRP159526 | PRKX | 5613 | RNAseq | 0.5707 | 0.2414 | |
| SRP193095 | PRKX | 5613 | RNAseq | 0.0229 | 0.9239 | |
| SRP219564 | PRKX | 5613 | RNAseq | -0.3914 | 0.5218 | |
| TCGA | PRKX | 5613 | RNAseq | 0.3759 | 0.0005 |
Upregulated datasets: 0; Downregulated datasets: 1.
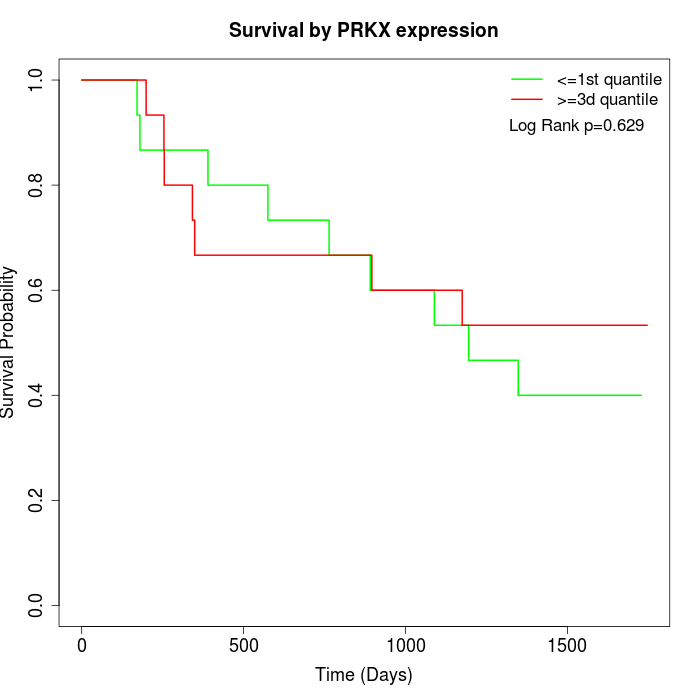
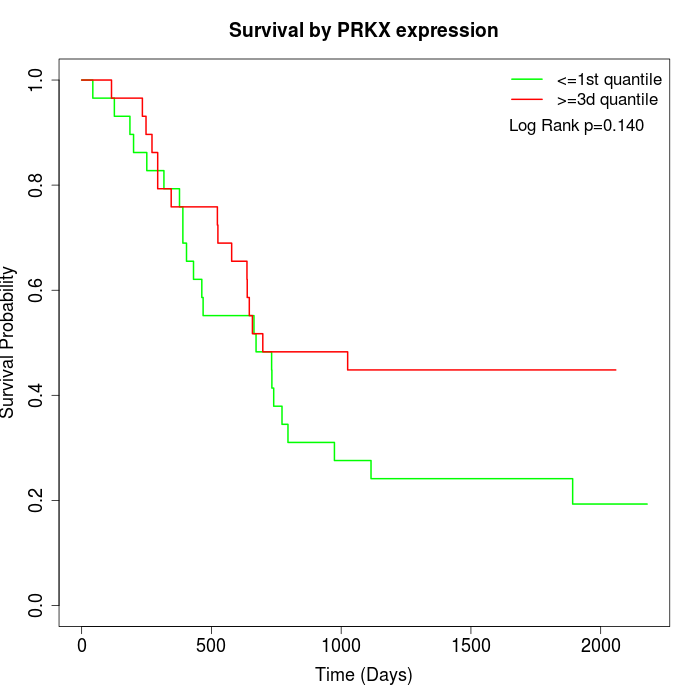
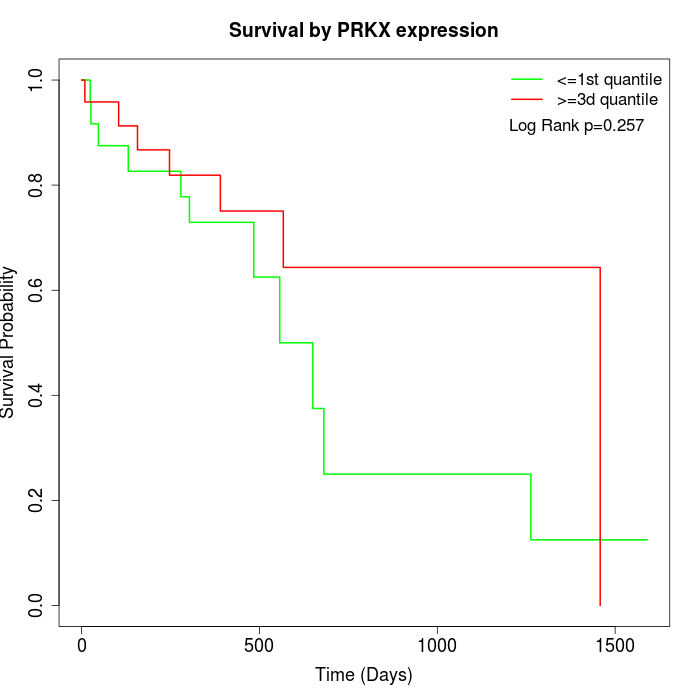
Survival by PRKX expression:
Note: Click image to view full size file.
Copy number change of PRKX:
No record found for this gene.
Somatic mutations of PRKX:
Generating mutation plots.
Highly correlated genes for PRKX:
Showing top 20/350 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PRKX | VMA21 | 0.769252 | 3 | 0 | 3 |
| PRKX | NUFIP2 | 0.744552 | 3 | 0 | 3 |
| PRKX | PEX2 | 0.741868 | 3 | 0 | 3 |
| PRKX | RNF34 | 0.737508 | 3 | 0 | 3 |
| PRKX | NCAPD2 | 0.728709 | 3 | 0 | 3 |
| PRKX | SEC61A1 | 0.72702 | 3 | 0 | 3 |
| PRKX | ZFYVE27 | 0.713697 | 3 | 0 | 3 |
| PRKX | ASNSD1 | 0.708318 | 3 | 0 | 3 |
| PRKX | HCLS1 | 0.705174 | 3 | 0 | 3 |
| PRKX | SRP9 | 0.703356 | 3 | 0 | 3 |
| PRKX | LSR | 0.699296 | 3 | 0 | 3 |
| PRKX | SLC35A2 | 0.696555 | 3 | 0 | 3 |
| PRKX | OSGEP | 0.692926 | 3 | 0 | 3 |
| PRKX | LINC01000 | 0.691425 | 3 | 0 | 3 |
| PRKX | MIA3 | 0.688866 | 4 | 0 | 3 |
| PRKX | LIN9 | 0.688701 | 3 | 0 | 3 |
| PRKX | QSOX2 | 0.685917 | 3 | 0 | 3 |
| PRKX | ACBD3 | 0.684785 | 3 | 0 | 3 |
| PRKX | MARS2 | 0.682496 | 4 | 0 | 3 |
| PRKX | CCDC117 | 0.682121 | 3 | 0 | 3 |
For details and further investigation, click here