| Full name: Prader-Willi region non-protein coding RNA 2 | Alias Symbol: NCRNA00199 | ||
| Type: non-coding RNA | Cytoband: 15q11.2 | ||
| Entrez ID: 791115 | HGNC ID: HGNC:33236 | Ensembl Gene: ENSG00000260551 | OMIM ID: 611217 |
| Drug and gene relationship at DGIdb | |||
Expression of PWRN2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PWRN2 | 791115 | 1568663_a_at | -0.0710 | 0.7296 | |
| GSE26886 | PWRN2 | 791115 | 1568663_a_at | 0.0879 | 0.4724 | |
| GSE45670 | PWRN2 | 791115 | 1568663_a_at | 0.0353 | 0.6471 | |
| GSE53622 | PWRN2 | 791115 | 7571 | 0.1048 | 0.4885 | |
| GSE53624 | PWRN2 | 791115 | 7571 | 0.3583 | 0.0008 | |
| GSE63941 | PWRN2 | 791115 | 1568663_a_at | 0.0960 | 0.5770 | |
| GSE77861 | PWRN2 | 791115 | 1568663_a_at | -0.0328 | 0.8018 | |
| TCGA | PWRN2 | 791115 | RNAseq | 1.6176 | 0.4608 |
Upregulated datasets: 0; Downregulated datasets: 0.
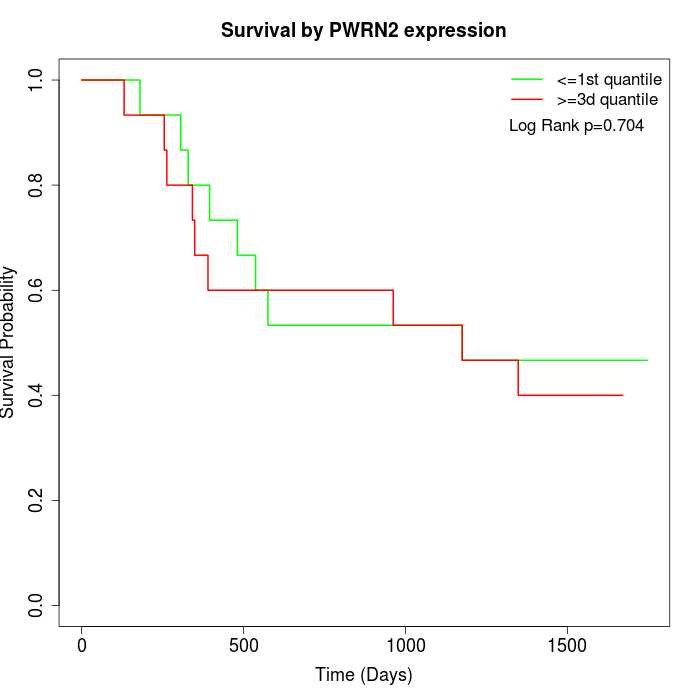
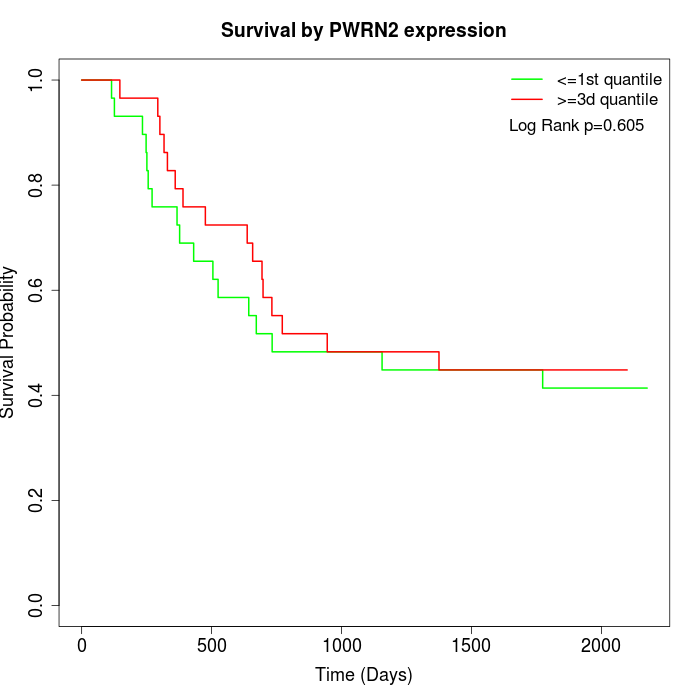
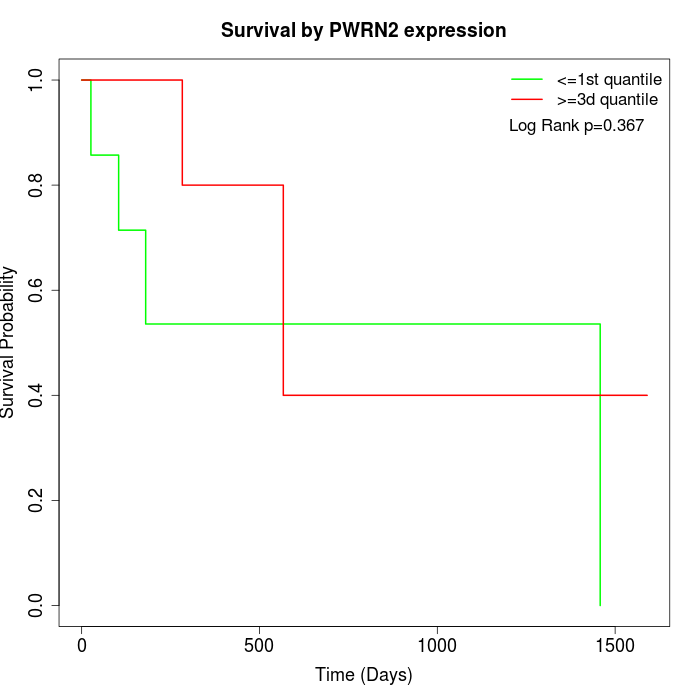
Survival by PWRN2 expression:
Note: Click image to view full size file.
Copy number change of PWRN2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PWRN2 | 791115 | 2 | 7 | 21 | |
| GSE20123 | PWRN2 | 791115 | 2 | 7 | 21 | |
| GSE43470 | PWRN2 | 791115 | 3 | 5 | 35 | |
| GSE46452 | PWRN2 | 791115 | 3 | 7 | 49 | |
| GSE47630 | PWRN2 | 791115 | 8 | 11 | 21 | |
| GSE54993 | PWRN2 | 791115 | 4 | 6 | 60 | |
| GSE54994 | PWRN2 | 791115 | 7 | 6 | 40 | |
| GSE60625 | PWRN2 | 791115 | 4 | 3 | 4 | |
| GSE74703 | PWRN2 | 791115 | 3 | 3 | 30 | |
| GSE74704 | PWRN2 | 791115 | 1 | 5 | 14 | |
| TCGA | PWRN2 | 791115 | 17 | 20 | 59 |
Total number of gains: 54; Total number of losses: 80; Total Number of normals: 354.
Somatic mutations of PWRN2:
Generating mutation plots.
Highly correlated genes for PWRN2:
Showing top 20/28 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PWRN2 | CNPY1 | 0.660961 | 3 | 0 | 3 |
| PWRN2 | LINC00857 | 0.640944 | 3 | 0 | 3 |
| PWRN2 | HCRT | 0.638886 | 3 | 0 | 3 |
| PWRN2 | FOXD4L5 | 0.630889 | 3 | 0 | 3 |
| PWRN2 | C1orf194 | 0.61448 | 3 | 0 | 3 |
| PWRN2 | SPAM1 | 0.612019 | 4 | 0 | 3 |
| PWRN2 | SIGLEC6 | 0.598149 | 4 | 0 | 3 |
| PWRN2 | CCDC38 | 0.581369 | 3 | 0 | 3 |
| PWRN2 | LINC00163 | 0.578227 | 4 | 0 | 3 |
| PWRN2 | KCNB2 | 0.576169 | 3 | 0 | 3 |
| PWRN2 | GFRA2 | 0.574573 | 4 | 0 | 3 |
| PWRN2 | TAC3 | 0.572259 | 4 | 0 | 3 |
| PWRN2 | TXNDC2 | 0.569384 | 4 | 0 | 3 |
| PWRN2 | MYL1 | 0.56552 | 4 | 0 | 3 |
| PWRN2 | KCNJ1 | 0.559904 | 4 | 0 | 3 |
| PWRN2 | SLC38A3 | 0.558244 | 3 | 0 | 3 |
| PWRN2 | MS4A6E | 0.554086 | 3 | 0 | 3 |
| PWRN2 | TMEM92 | 0.551616 | 4 | 0 | 3 |
| PWRN2 | TMEM207 | 0.549487 | 4 | 0 | 3 |
| PWRN2 | SPINT3 | 0.544247 | 6 | 0 | 3 |
For details and further investigation, click here