| Full name: RAD52 homolog, DNA repair protein | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 12p13.33 | ||
| Entrez ID: 5893 | HGNC ID: HGNC:9824 | Ensembl Gene: ENSG00000002016 | OMIM ID: 600392 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of RAD52:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RAD52 | 5893 | 236880_at | -0.1214 | 0.7908 | |
| GSE20347 | RAD52 | 5893 | 205647_at | 0.3680 | 0.0113 | |
| GSE23400 | RAD52 | 5893 | 211904_x_at | -0.1770 | 0.0001 | |
| GSE26886 | RAD52 | 5893 | 236880_at | 0.3920 | 0.0020 | |
| GSE29001 | RAD52 | 5893 | 205647_at | 0.4747 | 0.1749 | |
| GSE38129 | RAD52 | 5893 | 205647_at | 0.2733 | 0.0232 | |
| GSE45670 | RAD52 | 5893 | 236880_at | 0.1970 | 0.0604 | |
| GSE53622 | RAD52 | 5893 | 37080 | 0.3267 | 0.0000 | |
| GSE53624 | RAD52 | 5893 | 29827 | 0.5698 | 0.0000 | |
| GSE63941 | RAD52 | 5893 | 236880_at | 0.1085 | 0.5419 | |
| GSE77861 | RAD52 | 5893 | 236880_at | -0.1374 | 0.3836 | |
| GSE97050 | RAD52 | 5893 | A_33_P3242703 | 0.2198 | 0.3379 | |
| SRP007169 | RAD52 | 5893 | RNAseq | 0.3414 | 0.6211 | |
| SRP064894 | RAD52 | 5893 | RNAseq | 0.3954 | 0.0555 | |
| SRP133303 | RAD52 | 5893 | RNAseq | 0.1620 | 0.4931 | |
| SRP159526 | RAD52 | 5893 | RNAseq | 0.2367 | 0.5531 | |
| SRP193095 | RAD52 | 5893 | RNAseq | 0.2068 | 0.1581 | |
| SRP219564 | RAD52 | 5893 | RNAseq | 0.3338 | 0.0962 | |
| TCGA | RAD52 | 5893 | RNAseq | -0.0676 | 0.4482 |
Upregulated datasets: 0; Downregulated datasets: 0.
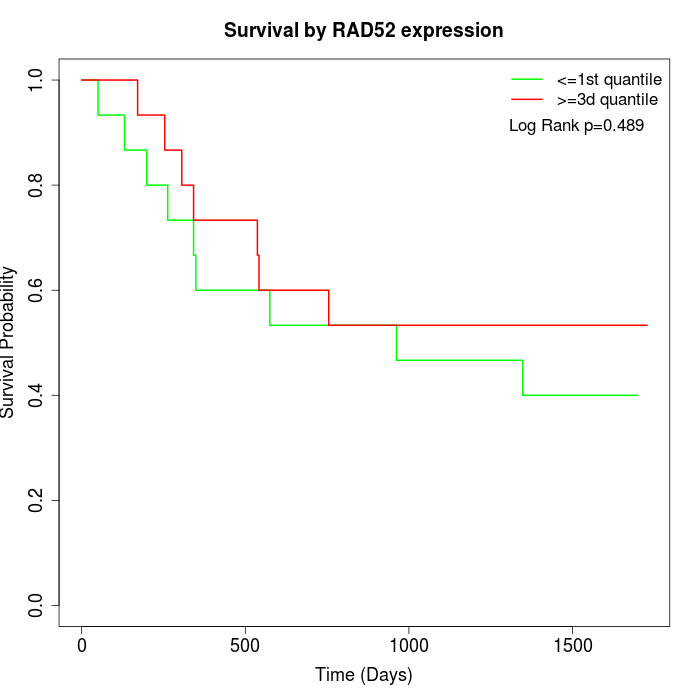
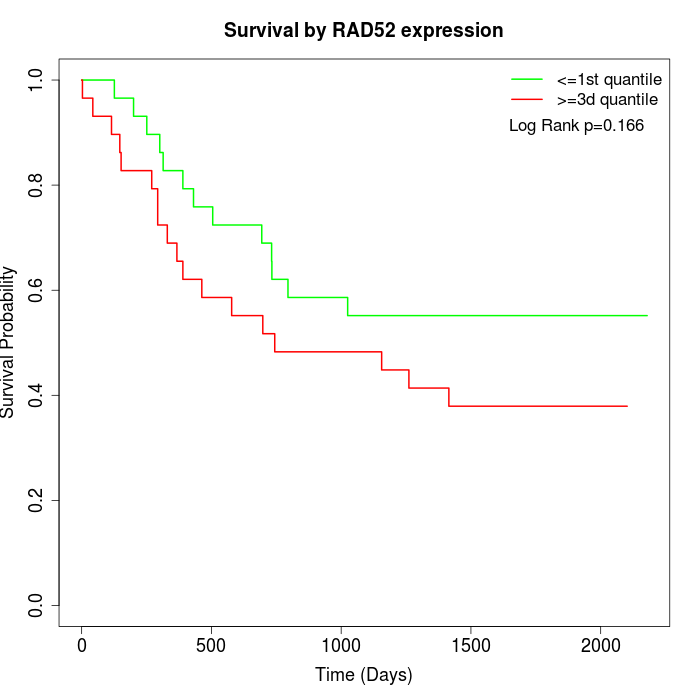
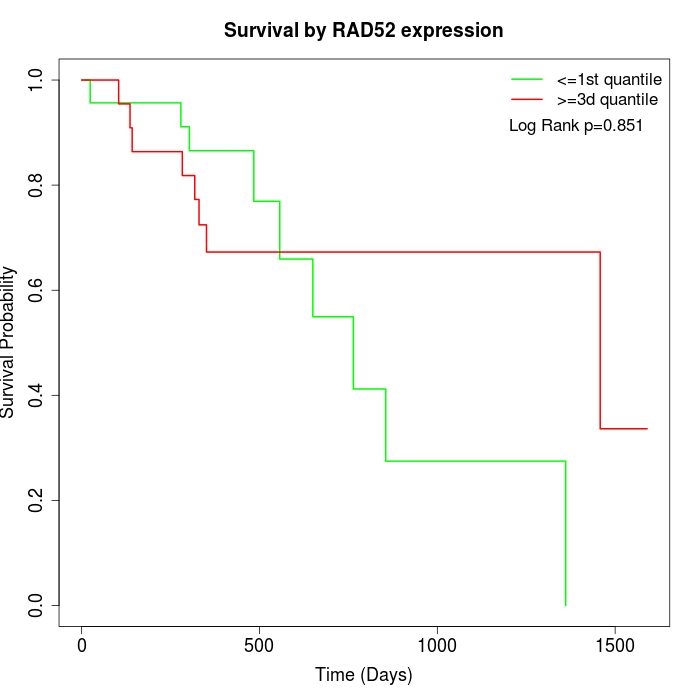
Survival by RAD52 expression:
Note: Click image to view full size file.
Copy number change of RAD52:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RAD52 | 5893 | 9 | 4 | 17 | |
| GSE20123 | RAD52 | 5893 | 8 | 4 | 18 | |
| GSE43470 | RAD52 | 5893 | 8 | 3 | 32 | |
| GSE46452 | RAD52 | 5893 | 10 | 1 | 48 | |
| GSE47630 | RAD52 | 5893 | 12 | 2 | 26 | |
| GSE54993 | RAD52 | 5893 | 1 | 10 | 59 | |
| GSE54994 | RAD52 | 5893 | 10 | 2 | 41 | |
| GSE60625 | RAD52 | 5893 | 0 | 1 | 10 | |
| GSE74703 | RAD52 | 5893 | 7 | 2 | 27 | |
| GSE74704 | RAD52 | 5893 | 5 | 2 | 13 | |
| TCGA | RAD52 | 5893 | 39 | 8 | 49 |
Total number of gains: 109; Total number of losses: 39; Total Number of normals: 340.
Somatic mutations of RAD52:
Generating mutation plots.
Highly correlated genes for RAD52:
Showing top 20/357 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RAD52 | ADM2 | 0.859089 | 3 | 0 | 3 |
| RAD52 | BEAN1 | 0.788033 | 3 | 0 | 3 |
| RAD52 | PSPN | 0.786895 | 3 | 0 | 3 |
| RAD52 | TULP1 | 0.762019 | 3 | 0 | 3 |
| RAD52 | OSBPL5 | 0.76133 | 3 | 0 | 3 |
| RAD52 | RIMS4 | 0.759587 | 3 | 0 | 3 |
| RAD52 | MYO1A | 0.755265 | 3 | 0 | 3 |
| RAD52 | PLA2G4F | 0.751776 | 3 | 0 | 3 |
| RAD52 | BTNL2 | 0.749226 | 3 | 0 | 3 |
| RAD52 | SLC6A18 | 0.748488 | 3 | 0 | 3 |
| RAD52 | MGAT5B | 0.739858 | 4 | 0 | 4 |
| RAD52 | NAT8B | 0.739522 | 3 | 0 | 3 |
| RAD52 | NKX2-8 | 0.739056 | 4 | 0 | 4 |
| RAD52 | MAG | 0.738986 | 4 | 0 | 4 |
| RAD52 | BMP10 | 0.738864 | 4 | 0 | 4 |
| RAD52 | TAS2R30 | 0.736655 | 4 | 0 | 4 |
| RAD52 | ZNF219 | 0.736501 | 3 | 0 | 3 |
| RAD52 | SHANK1 | 0.734675 | 3 | 0 | 3 |
| RAD52 | VEPH1 | 0.728563 | 4 | 0 | 4 |
| RAD52 | CACNA1I | 0.72571 | 3 | 0 | 3 |
For details and further investigation, click here