| Full name: RAS p21 protein activator 1 | Alias Symbol: GAP|CM-AVM|p120GAP|p120RASGAP|p120 | ||
| Type: protein-coding gene | Cytoband: 5q14.3 | ||
| Entrez ID: 5921 | HGNC ID: HGNC:9871 | Ensembl Gene: ENSG00000145715 | OMIM ID: 139150 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
RASA1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04010 | MAPK signaling pathway | |
| hsa04014 | Ras signaling pathway | |
| hsa04360 | Axon guidance |
Expression of RASA1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RASA1 | 5921 | 202677_at | -0.2575 | 0.5089 | |
| GSE20347 | RASA1 | 5921 | 202677_at | -0.5558 | 0.0040 | |
| GSE23400 | RASA1 | 5921 | 202677_at | -0.1611 | 0.0587 | |
| GSE26886 | RASA1 | 5921 | 202677_at | -0.2374 | 0.2834 | |
| GSE29001 | RASA1 | 5921 | 202677_at | -0.1350 | 0.6505 | |
| GSE38129 | RASA1 | 5921 | 202677_at | -0.2995 | 0.0433 | |
| GSE45670 | RASA1 | 5921 | 202677_at | -0.0164 | 0.9365 | |
| GSE53622 | RASA1 | 5921 | 69475 | -0.1235 | 0.0419 | |
| GSE53624 | RASA1 | 5921 | 52456 | -0.3236 | 0.0000 | |
| GSE63941 | RASA1 | 5921 | 202677_at | -0.6047 | 0.2101 | |
| GSE77861 | RASA1 | 5921 | 202677_at | -0.7026 | 0.1846 | |
| GSE97050 | RASA1 | 5921 | A_23_P18939 | 0.2843 | 0.2665 | |
| SRP007169 | RASA1 | 5921 | RNAseq | 0.0873 | 0.8112 | |
| SRP008496 | RASA1 | 5921 | RNAseq | -0.3130 | 0.1703 | |
| SRP064894 | RASA1 | 5921 | RNAseq | -0.1127 | 0.6479 | |
| SRP133303 | RASA1 | 5921 | RNAseq | 0.0264 | 0.8844 | |
| SRP159526 | RASA1 | 5921 | RNAseq | -0.6748 | 0.0259 | |
| SRP193095 | RASA1 | 5921 | RNAseq | -0.3797 | 0.0000 | |
| SRP219564 | RASA1 | 5921 | RNAseq | -0.5896 | 0.0941 | |
| TCGA | RASA1 | 5921 | RNAseq | 0.0024 | 0.9671 |
Upregulated datasets: 0; Downregulated datasets: 0.
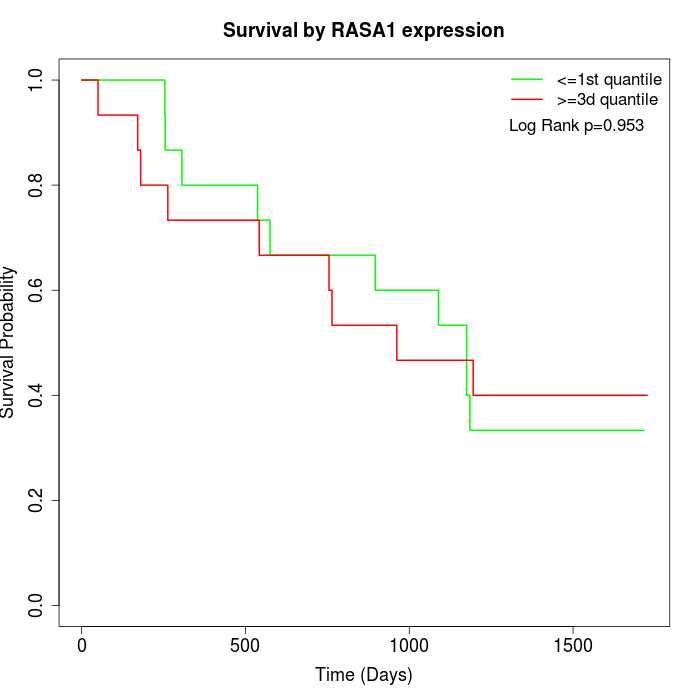
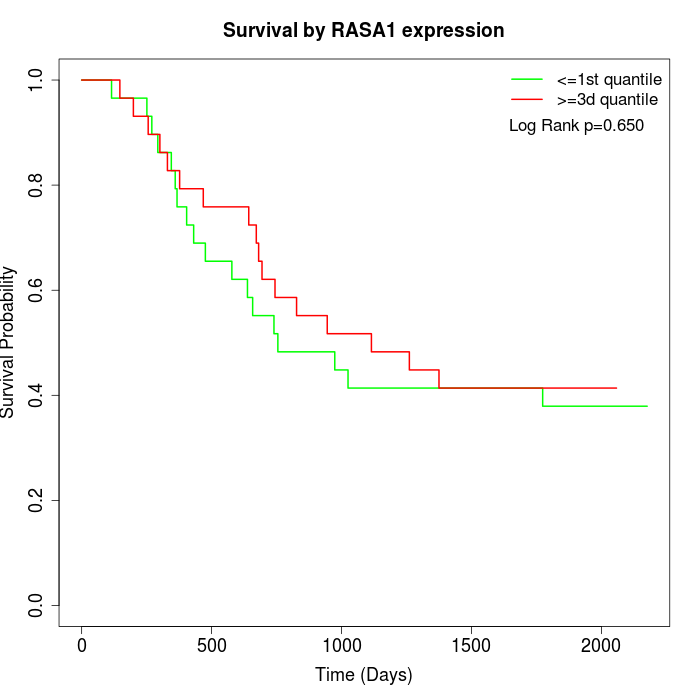
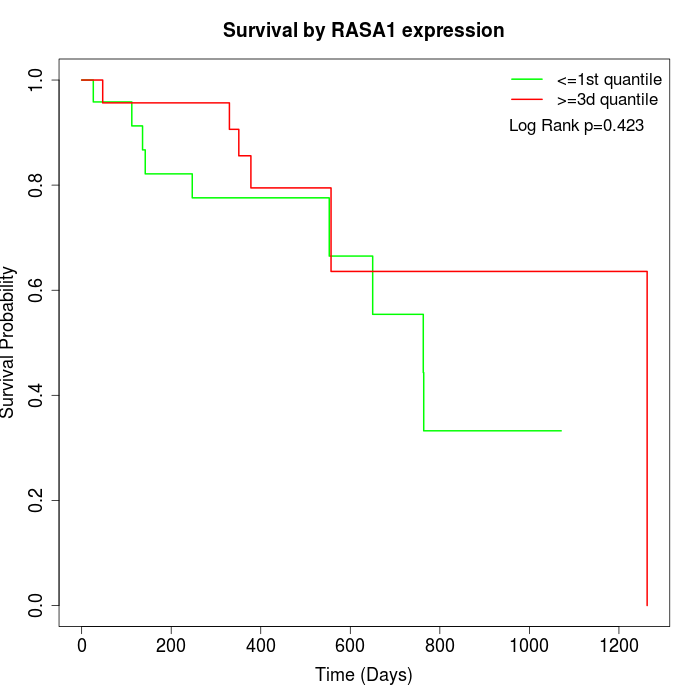
Survival by RASA1 expression:
Note: Click image to view full size file.
Copy number change of RASA1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RASA1 | 5921 | 1 | 14 | 15 | |
| GSE20123 | RASA1 | 5921 | 1 | 14 | 15 | |
| GSE43470 | RASA1 | 5921 | 2 | 10 | 31 | |
| GSE46452 | RASA1 | 5921 | 0 | 28 | 31 | |
| GSE47630 | RASA1 | 5921 | 1 | 20 | 19 | |
| GSE54993 | RASA1 | 5921 | 8 | 1 | 61 | |
| GSE54994 | RASA1 | 5921 | 1 | 18 | 34 | |
| GSE60625 | RASA1 | 5921 | 0 | 0 | 11 | |
| GSE74703 | RASA1 | 5921 | 1 | 8 | 27 | |
| GSE74704 | RASA1 | 5921 | 1 | 7 | 12 | |
| TCGA | RASA1 | 5921 | 2 | 51 | 43 |
Total number of gains: 18; Total number of losses: 171; Total Number of normals: 299.
Somatic mutations of RASA1:
Generating mutation plots.
Highly correlated genes for RASA1:
Showing top 20/533 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RASA1 | CHMP4B | 0.804263 | 3 | 0 | 3 |
| RASA1 | SAMD8 | 0.754367 | 3 | 0 | 3 |
| RASA1 | ANGPTL4 | 0.751381 | 3 | 0 | 3 |
| RASA1 | BLOC1S2 | 0.745121 | 3 | 0 | 3 |
| RASA1 | RSPRY1 | 0.743399 | 3 | 0 | 3 |
| RASA1 | ATG2B | 0.742315 | 4 | 0 | 4 |
| RASA1 | TINF2 | 0.729761 | 3 | 0 | 3 |
| RASA1 | TMEM161B | 0.727885 | 4 | 0 | 4 |
| RASA1 | TTC39B | 0.697621 | 3 | 0 | 3 |
| RASA1 | MLKL | 0.694158 | 3 | 0 | 3 |
| RASA1 | TMEM19 | 0.693801 | 6 | 0 | 5 |
| RASA1 | PSMD7 | 0.691674 | 4 | 0 | 3 |
| RASA1 | KCTD18 | 0.691281 | 4 | 0 | 3 |
| RASA1 | HUS1 | 0.691035 | 4 | 0 | 3 |
| RASA1 | ZBTB43 | 0.69089 | 3 | 0 | 3 |
| RASA1 | MPP6 | 0.685292 | 3 | 0 | 3 |
| RASA1 | SUSD1 | 0.68351 | 4 | 0 | 3 |
| RASA1 | ZNF441 | 0.681488 | 3 | 0 | 3 |
| RASA1 | HSPA4 | 0.679654 | 5 | 0 | 4 |
| RASA1 | TMEM120A | 0.676735 | 3 | 0 | 3 |
For details and further investigation, click here