| Full name: RNA binding motif single stranded interacting protein 1 | Alias Symbol: SCR2|MSSP-1|MSSP-2|MSSP-3|YC1|HCC-4|DKFZp564H0764 | ||
| Type: protein-coding gene | Cytoband: 2q24.2 | ||
| Entrez ID: 5937 | HGNC ID: HGNC:9907 | Ensembl Gene: ENSG00000153250 | OMIM ID: 602310 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of RBMS1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RBMS1 | 5937 | 215127_s_at | 0.3516 | 0.5169 | |
| GSE20347 | RBMS1 | 5937 | 215127_s_at | 0.4038 | 0.0094 | |
| GSE23400 | RBMS1 | 5937 | 203748_x_at | 0.0850 | 0.2200 | |
| GSE26886 | RBMS1 | 5937 | 225265_at | -0.4893 | 0.0537 | |
| GSE29001 | RBMS1 | 5937 | 215127_s_at | 0.3198 | 0.2474 | |
| GSE38129 | RBMS1 | 5937 | 215127_s_at | 0.2579 | 0.0543 | |
| GSE45670 | RBMS1 | 5937 | 215127_s_at | -0.0950 | 0.5582 | |
| GSE53622 | RBMS1 | 5937 | 51956 | 0.2616 | 0.0001 | |
| GSE53624 | RBMS1 | 5937 | 51956 | 0.5466 | 0.0000 | |
| GSE63941 | RBMS1 | 5937 | 215127_s_at | -0.9861 | 0.0208 | |
| GSE77861 | RBMS1 | 5937 | 215127_s_at | 0.5040 | 0.1023 | |
| GSE97050 | RBMS1 | 5937 | A_23_P431410 | 0.0591 | 0.8066 | |
| SRP007169 | RBMS1 | 5937 | RNAseq | -0.6410 | 0.1582 | |
| SRP008496 | RBMS1 | 5937 | RNAseq | -0.4452 | 0.0946 | |
| SRP064894 | RBMS1 | 5937 | RNAseq | 0.1144 | 0.5620 | |
| SRP133303 | RBMS1 | 5937 | RNAseq | 0.5068 | 0.0112 | |
| SRP159526 | RBMS1 | 5937 | RNAseq | 0.1892 | 0.3476 | |
| SRP193095 | RBMS1 | 5937 | RNAseq | 0.1661 | 0.2393 | |
| SRP219564 | RBMS1 | 5937 | RNAseq | 0.0499 | 0.9048 | |
| TCGA | RBMS1 | 5937 | RNAseq | 0.1226 | 0.0135 |
Upregulated datasets: 0; Downregulated datasets: 0.
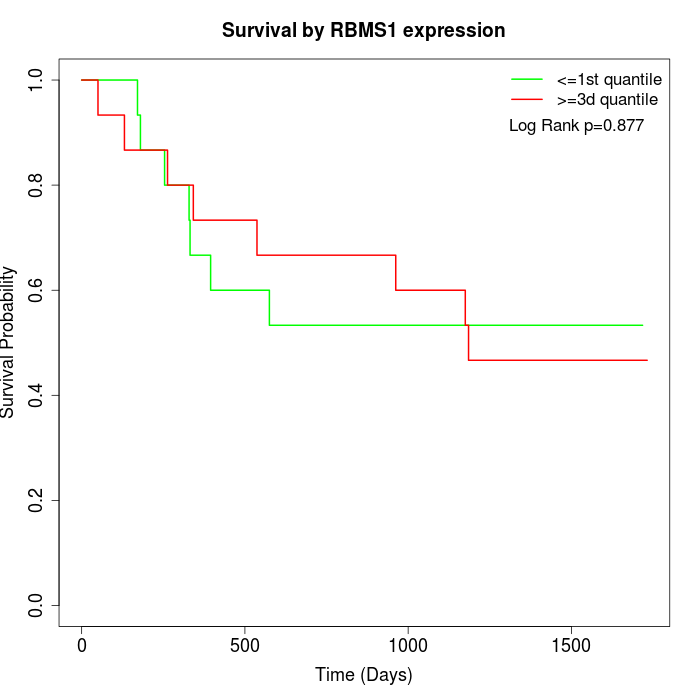
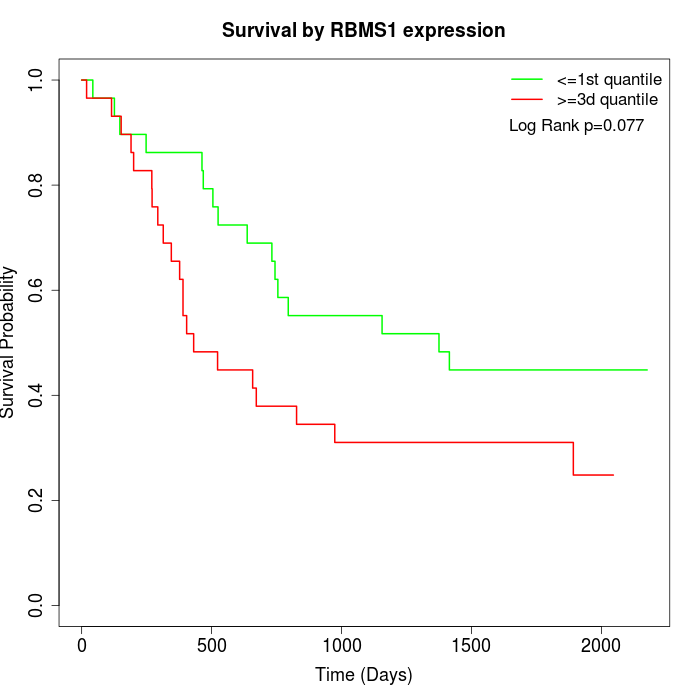
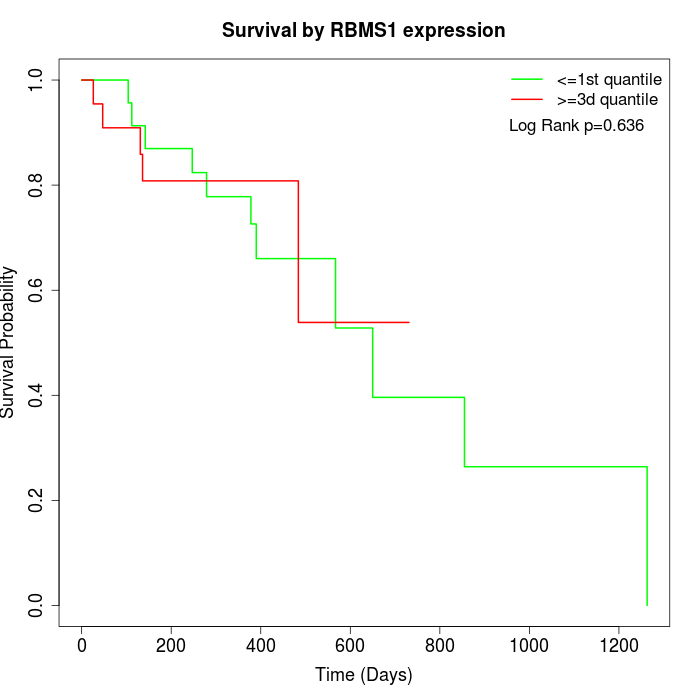
Survival by RBMS1 expression:
Note: Click image to view full size file.
Copy number change of RBMS1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RBMS1 | 5937 | 5 | 1 | 24 | |
| GSE20123 | RBMS1 | 5937 | 5 | 1 | 24 | |
| GSE43470 | RBMS1 | 5937 | 4 | 1 | 38 | |
| GSE46452 | RBMS1 | 5937 | 1 | 4 | 54 | |
| GSE47630 | RBMS1 | 5937 | 5 | 3 | 32 | |
| GSE54993 | RBMS1 | 5937 | 0 | 4 | 66 | |
| GSE54994 | RBMS1 | 5937 | 11 | 2 | 40 | |
| GSE60625 | RBMS1 | 5937 | 0 | 3 | 8 | |
| GSE74703 | RBMS1 | 5937 | 3 | 1 | 32 | |
| GSE74704 | RBMS1 | 5937 | 3 | 0 | 17 | |
| TCGA | RBMS1 | 5937 | 23 | 10 | 63 |
Total number of gains: 60; Total number of losses: 30; Total Number of normals: 398.
Somatic mutations of RBMS1:
Generating mutation plots.
Highly correlated genes for RBMS1:
Showing top 20/241 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RBMS1 | PDHX | 0.816227 | 3 | 0 | 3 |
| RBMS1 | CWC22 | 0.782766 | 3 | 0 | 3 |
| RBMS1 | CEBPZ | 0.774852 | 4 | 0 | 4 |
| RBMS1 | UBXN2A | 0.75904 | 3 | 0 | 3 |
| RBMS1 | NUP35 | 0.751561 | 3 | 0 | 3 |
| RBMS1 | DDX59 | 0.733328 | 3 | 0 | 3 |
| RBMS1 | SREBF1 | 0.725745 | 3 | 0 | 3 |
| RBMS1 | EXOC8 | 0.725332 | 4 | 0 | 3 |
| RBMS1 | CCP110 | 0.722875 | 3 | 0 | 3 |
| RBMS1 | ZFYVE26 | 0.708775 | 3 | 0 | 3 |
| RBMS1 | KCTD21 | 0.707928 | 3 | 0 | 3 |
| RBMS1 | ZNF638 | 0.706151 | 4 | 0 | 4 |
| RBMS1 | UBE2K | 0.685079 | 3 | 0 | 3 |
| RBMS1 | ZNF23 | 0.675958 | 4 | 0 | 3 |
| RBMS1 | SEC16A | 0.672566 | 3 | 0 | 3 |
| RBMS1 | ADAP2 | 0.671702 | 3 | 0 | 3 |
| RBMS1 | FAF2 | 0.66921 | 3 | 0 | 3 |
| RBMS1 | DYNC1I2 | 0.66753 | 6 | 0 | 4 |
| RBMS1 | ANKRD36B | 0.666824 | 3 | 0 | 3 |
| RBMS1 | UBE2E2 | 0.666596 | 3 | 0 | 3 |
For details and further investigation, click here