| Full name: RNA binding motif protein X-linked 2 | Alias Symbol: CGI-79|Snu17 | ||
| Type: protein-coding gene | Cytoband: Xq26.1 | ||
| Entrez ID: 51634 | HGNC ID: HGNC:24282 | Ensembl Gene: ENSG00000134597 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of RBMX2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RBMX2 | 51634 | 204098_at | -0.1249 | 0.6998 | |
| GSE20347 | RBMX2 | 51634 | 204098_at | 0.4546 | 0.0000 | |
| GSE23400 | RBMX2 | 51634 | 204098_at | 0.2423 | 0.0000 | |
| GSE26886 | RBMX2 | 51634 | 204098_at | 0.5327 | 0.0001 | |
| GSE29001 | RBMX2 | 51634 | 204098_at | 0.5071 | 0.0158 | |
| GSE38129 | RBMX2 | 51634 | 204098_at | 0.4191 | 0.0000 | |
| GSE45670 | RBMX2 | 51634 | 204098_at | 0.3088 | 0.0275 | |
| GSE53622 | RBMX2 | 51634 | 26825 | 0.2191 | 0.0003 | |
| GSE53624 | RBMX2 | 51634 | 26825 | 0.3788 | 0.0000 | |
| GSE63941 | RBMX2 | 51634 | 204098_at | -0.0169 | 0.9676 | |
| GSE77861 | RBMX2 | 51634 | 204098_at | 0.6209 | 0.0014 | |
| GSE97050 | RBMX2 | 51634 | A_23_P255153 | -0.1350 | 0.5329 | |
| SRP007169 | RBMX2 | 51634 | RNAseq | 0.1232 | 0.7705 | |
| SRP008496 | RBMX2 | 51634 | RNAseq | 0.2181 | 0.3452 | |
| SRP064894 | RBMX2 | 51634 | RNAseq | 0.5785 | 0.0485 | |
| SRP133303 | RBMX2 | 51634 | RNAseq | 0.6905 | 0.0003 | |
| SRP159526 | RBMX2 | 51634 | RNAseq | 0.3625 | 0.0560 | |
| SRP193095 | RBMX2 | 51634 | RNAseq | 0.1625 | 0.1915 | |
| SRP219564 | RBMX2 | 51634 | RNAseq | 0.0502 | 0.8576 | |
| TCGA | RBMX2 | 51634 | RNAseq | 0.1600 | 0.0070 |
Upregulated datasets: 0; Downregulated datasets: 0.
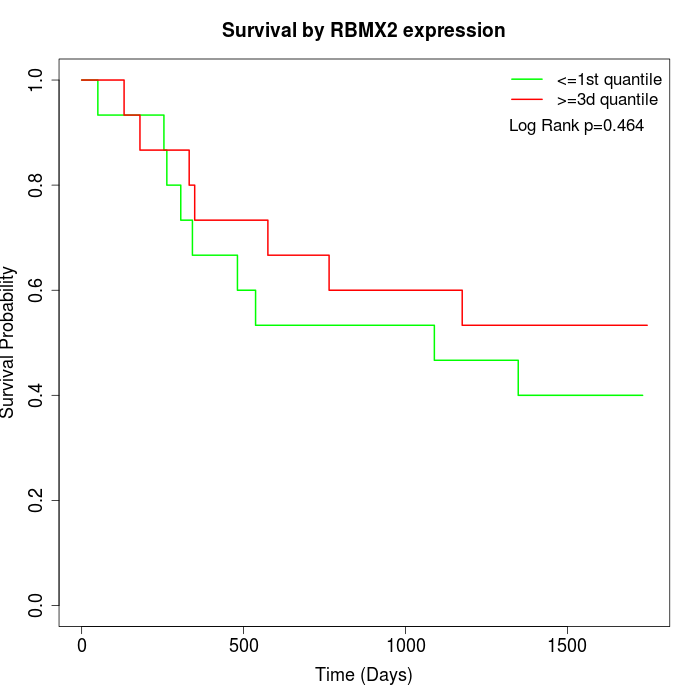
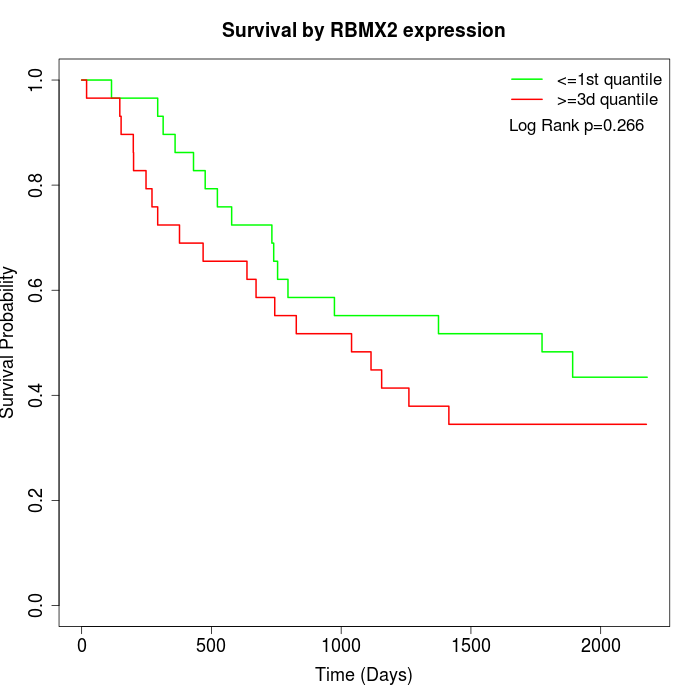
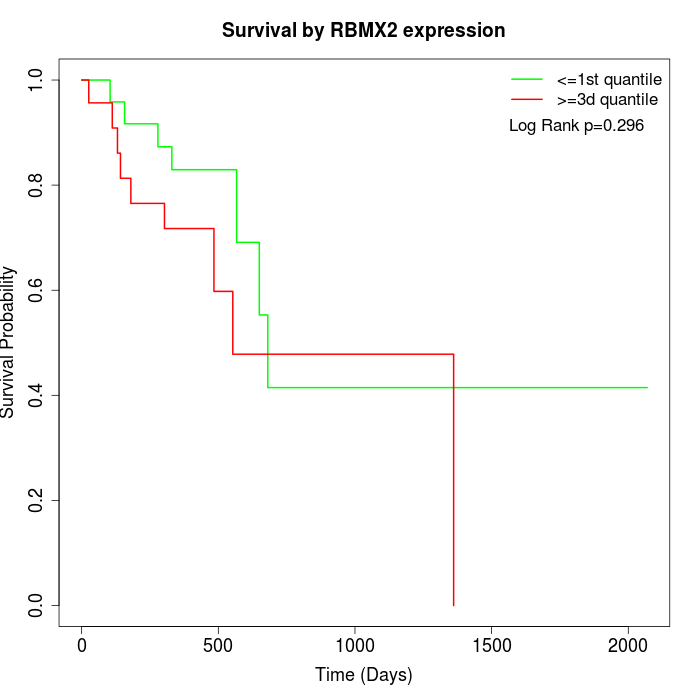
Survival by RBMX2 expression:
Note: Click image to view full size file.
Copy number change of RBMX2:
No record found for this gene.
Somatic mutations of RBMX2:
Generating mutation plots.
Highly correlated genes for RBMX2:
Showing top 20/1242 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RBMX2 | CLIP3 | 0.872605 | 3 | 0 | 3 |
| RBMX2 | UBR1 | 0.811516 | 3 | 0 | 3 |
| RBMX2 | MMRN2 | 0.790055 | 3 | 0 | 3 |
| RBMX2 | ZBTB26 | 0.784822 | 3 | 0 | 3 |
| RBMX2 | FAM76B | 0.783236 | 3 | 0 | 3 |
| RBMX2 | HABP4 | 0.777981 | 3 | 0 | 3 |
| RBMX2 | MTRF1L | 0.776623 | 3 | 0 | 3 |
| RBMX2 | FGF2 | 0.775761 | 3 | 0 | 3 |
| RBMX2 | PET117 | 0.775052 | 3 | 0 | 3 |
| RBMX2 | SOBP | 0.774015 | 3 | 0 | 3 |
| RBMX2 | CEP95 | 0.76785 | 3 | 0 | 3 |
| RBMX2 | CORO2B | 0.763072 | 3 | 0 | 3 |
| RBMX2 | SEMA3A | 0.762295 | 3 | 0 | 3 |
| RBMX2 | SPRED2 | 0.760848 | 3 | 0 | 3 |
| RBMX2 | ZNF417 | 0.760633 | 3 | 0 | 3 |
| RBMX2 | PIGU | 0.757787 | 3 | 0 | 3 |
| RBMX2 | CPT1A | 0.755545 | 3 | 0 | 3 |
| RBMX2 | IRF2BP2 | 0.746548 | 3 | 0 | 3 |
| RBMX2 | TSC22D1 | 0.745036 | 4 | 0 | 4 |
| RBMX2 | SETBP1 | 0.742016 | 3 | 0 | 3 |
For details and further investigation, click here