| Full name: LAS1 like, ribosome biogenesis factor | Alias Symbol: FLJ12525 | ||
| Type: protein-coding gene | Cytoband: Xq12 | ||
| Entrez ID: 81887 | HGNC ID: HGNC:25726 | Ensembl Gene: ENSG00000001497 | OMIM ID: 300964 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of LAS1L:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LAS1L | 81887 | 208117_s_at | 0.4249 | 0.1153 | |
| GSE20347 | LAS1L | 81887 | 208117_s_at | 0.3300 | 0.0244 | |
| GSE23400 | LAS1L | 81887 | 208117_s_at | 0.2796 | 0.0000 | |
| GSE26886 | LAS1L | 81887 | 208117_s_at | 0.0084 | 0.9794 | |
| GSE29001 | LAS1L | 81887 | 208117_s_at | 0.3290 | 0.1428 | |
| GSE38129 | LAS1L | 81887 | 208117_s_at | 0.3390 | 0.0009 | |
| GSE45670 | LAS1L | 81887 | 208117_s_at | 0.2899 | 0.0254 | |
| GSE53622 | LAS1L | 81887 | 59825 | -0.5025 | 0.0000 | |
| GSE53624 | LAS1L | 81887 | 59825 | -0.2025 | 0.0089 | |
| GSE63941 | LAS1L | 81887 | 208117_s_at | 1.1800 | 0.0036 | |
| GSE77861 | LAS1L | 81887 | 208117_s_at | 0.3487 | 0.0847 | |
| GSE97050 | LAS1L | 81887 | A_23_P256021 | -0.4104 | 0.1983 | |
| SRP007169 | LAS1L | 81887 | RNAseq | 0.1576 | 0.7722 | |
| SRP008496 | LAS1L | 81887 | RNAseq | 0.1756 | 0.6272 | |
| SRP064894 | LAS1L | 81887 | RNAseq | 0.5099 | 0.0019 | |
| SRP133303 | LAS1L | 81887 | RNAseq | 0.4341 | 0.0830 | |
| SRP159526 | LAS1L | 81887 | RNAseq | 0.2112 | 0.2270 | |
| SRP193095 | LAS1L | 81887 | RNAseq | 0.3633 | 0.0037 | |
| SRP219564 | LAS1L | 81887 | RNAseq | 0.4710 | 0.0699 | |
| TCGA | LAS1L | 81887 | RNAseq | 0.1856 | 0.0005 |
Upregulated datasets: 1; Downregulated datasets: 0.
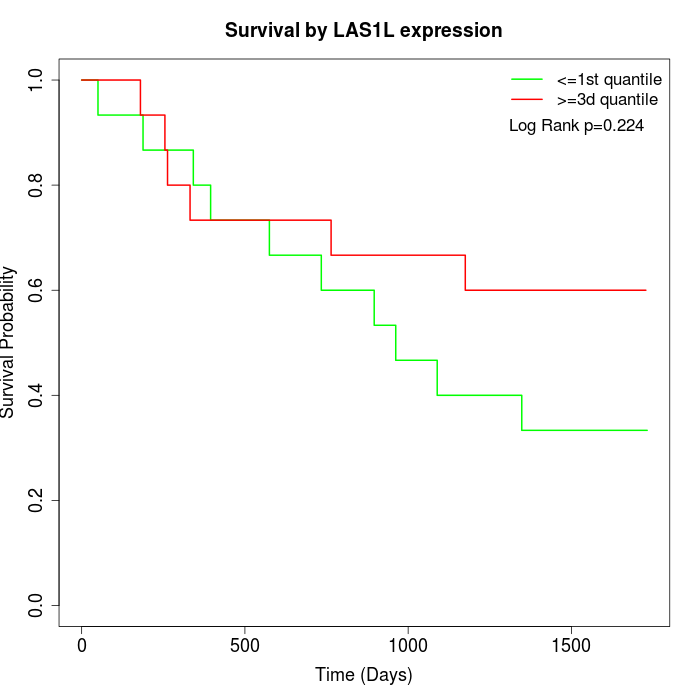
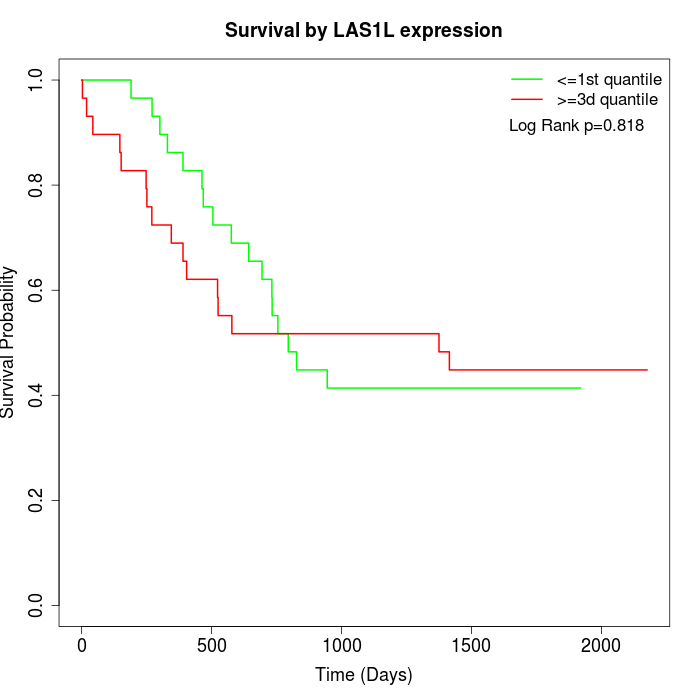
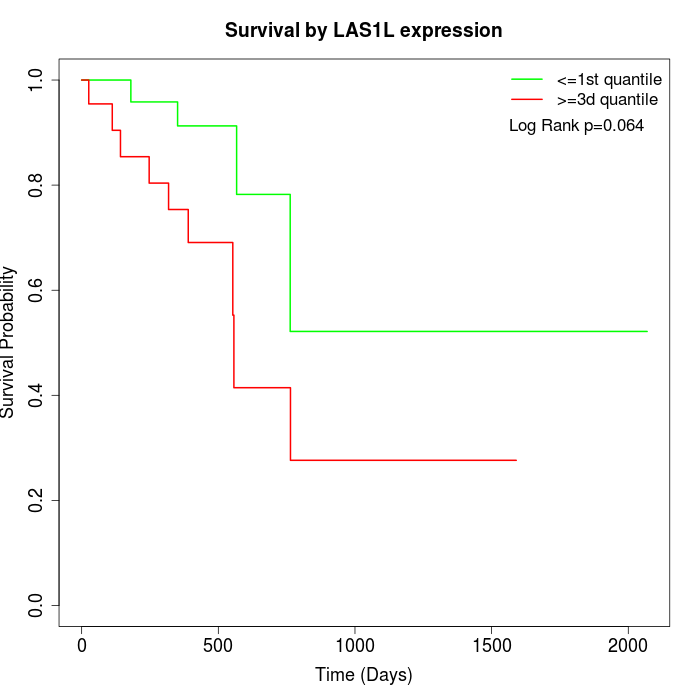
Survival by LAS1L expression:
Note: Click image to view full size file.
Copy number change of LAS1L:
No record found for this gene.
Somatic mutations of LAS1L:
Generating mutation plots.
Highly correlated genes for LAS1L:
Showing top 20/175 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LAS1L | FABP6 | 0.76875 | 3 | 0 | 3 |
| LAS1L | REM2 | 0.742075 | 3 | 0 | 3 |
| LAS1L | GPRIN1 | 0.669682 | 3 | 0 | 3 |
| LAS1L | MEGF10 | 0.668784 | 4 | 0 | 3 |
| LAS1L | CSTL1 | 0.668479 | 4 | 0 | 3 |
| LAS1L | ACOT8 | 0.664244 | 4 | 0 | 3 |
| LAS1L | NUP98 | 0.662598 | 3 | 0 | 3 |
| LAS1L | PPFIA3 | 0.657229 | 3 | 0 | 3 |
| LAS1L | CDK16 | 0.655458 | 3 | 0 | 3 |
| LAS1L | TCTE1 | 0.653917 | 3 | 0 | 3 |
| LAS1L | BEND3 | 0.650259 | 5 | 0 | 3 |
| LAS1L | TMEM68 | 0.649851 | 4 | 0 | 3 |
| LAS1L | MEI1 | 0.649711 | 3 | 0 | 3 |
| LAS1L | PRR5 | 0.643084 | 7 | 0 | 6 |
| LAS1L | MRPS17 | 0.639079 | 7 | 0 | 6 |
| LAS1L | POLR2F | 0.63636 | 5 | 0 | 5 |
| LAS1L | CCT7 | 0.635162 | 6 | 0 | 6 |
| LAS1L | HOTAIR | 0.633973 | 4 | 0 | 3 |
| LAS1L | ARID5A | 0.627984 | 3 | 0 | 3 |
| LAS1L | DNLZ | 0.627887 | 3 | 0 | 3 |
For details and further investigation, click here