| Full name: Ras related GTP binding B | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: Xp11.21 | ||
| Entrez ID: 10325 | HGNC ID: HGNC:19901 | Ensembl Gene: ENSG00000083750 | OMIM ID: 300725 |
| Drug and gene relationship at DGIdb | |||
RRAGB involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04150 | mTOR signaling pathway |
Expression of RRAGB:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RRAGB | 10325 | 205540_s_at | -0.2562 | 0.7883 | |
| GSE20347 | RRAGB | 10325 | 205540_s_at | -0.0753 | 0.2644 | |
| GSE23400 | RRAGB | 10325 | 205540_s_at | 0.0779 | 0.0693 | |
| GSE26886 | RRAGB | 10325 | 205540_s_at | -0.0095 | 0.9717 | |
| GSE29001 | RRAGB | 10325 | 205540_s_at | -0.0367 | 0.8619 | |
| GSE38129 | RRAGB | 10325 | 205540_s_at | -0.1540 | 0.4159 | |
| GSE45670 | RRAGB | 10325 | 205540_s_at | -0.2805 | 0.2182 | |
| GSE53622 | RRAGB | 10325 | 154038 | -0.1887 | 0.1041 | |
| GSE53624 | RRAGB | 10325 | 154038 | -0.1702 | 0.2862 | |
| GSE63941 | RRAGB | 10325 | 205540_s_at | -2.1975 | 0.0006 | |
| GSE77861 | RRAGB | 10325 | 205540_s_at | 0.0095 | 0.9371 | |
| GSE97050 | RRAGB | 10325 | A_23_P73721 | -0.0156 | 0.9509 | |
| SRP007169 | RRAGB | 10325 | RNAseq | 0.1991 | 0.6605 | |
| SRP008496 | RRAGB | 10325 | RNAseq | 0.8295 | 0.0470 | |
| SRP064894 | RRAGB | 10325 | RNAseq | 0.1216 | 0.4991 | |
| SRP133303 | RRAGB | 10325 | RNAseq | 0.0889 | 0.7045 | |
| SRP159526 | RRAGB | 10325 | RNAseq | 0.3826 | 0.0234 | |
| SRP193095 | RRAGB | 10325 | RNAseq | 0.0130 | 0.9080 | |
| SRP219564 | RRAGB | 10325 | RNAseq | -0.1387 | 0.7676 | |
| TCGA | RRAGB | 10325 | RNAseq | -0.1136 | 0.1564 |
Upregulated datasets: 0; Downregulated datasets: 1.
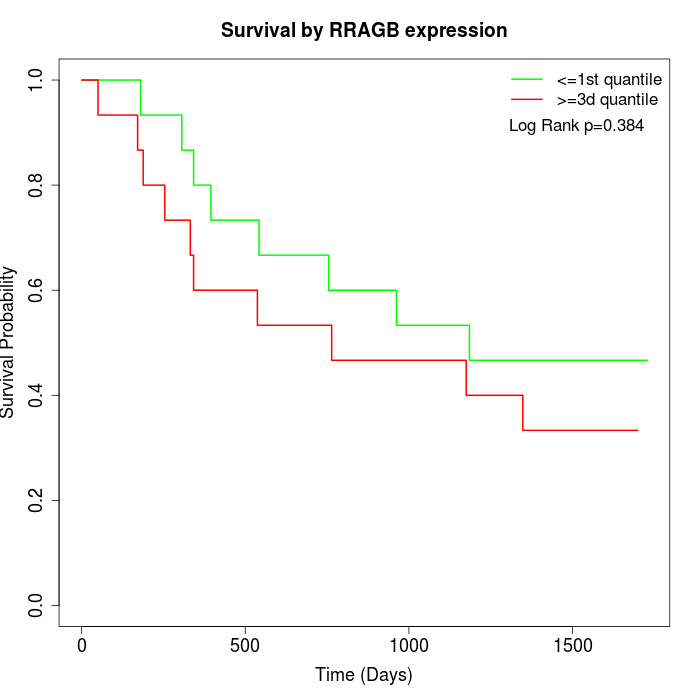
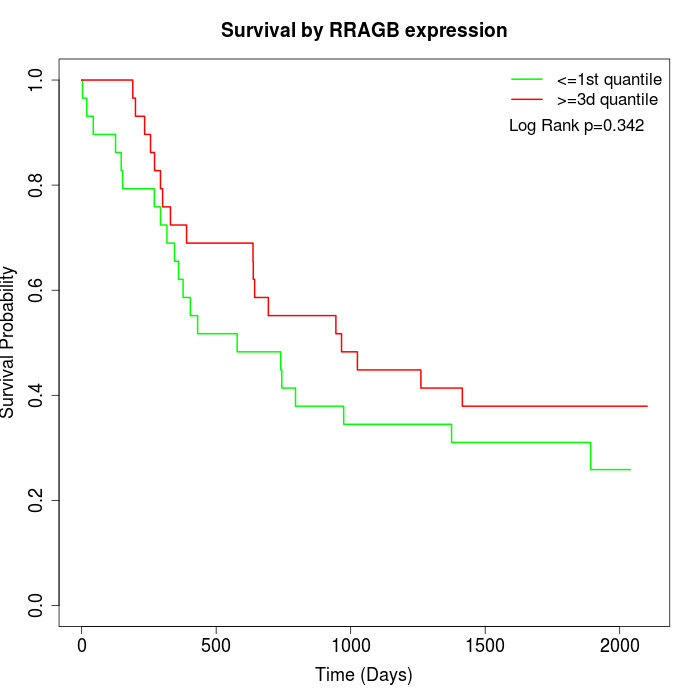
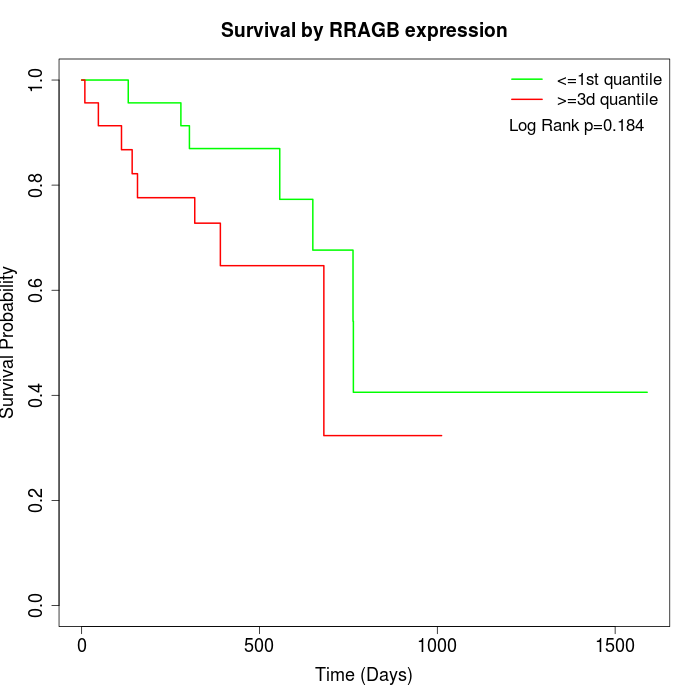
Survival by RRAGB expression:
Note: Click image to view full size file.
Copy number change of RRAGB:
No record found for this gene.
Somatic mutations of RRAGB:
Generating mutation plots.
Highly correlated genes for RRAGB:
Showing top 20/672 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RRAGB | LMBRD2 | 0.851618 | 3 | 0 | 3 |
| RRAGB | ZNF578 | 0.791067 | 3 | 0 | 3 |
| RRAGB | XBP1 | 0.767683 | 3 | 0 | 3 |
| RRAGB | PARD3B | 0.765466 | 3 | 0 | 3 |
| RRAGB | PNKD | 0.764273 | 3 | 0 | 3 |
| RRAGB | SCAF8 | 0.763449 | 3 | 0 | 3 |
| RRAGB | ZNF844 | 0.75633 | 3 | 0 | 3 |
| RRAGB | AFAP1 | 0.756204 | 3 | 0 | 3 |
| RRAGB | PARP16 | 0.755639 | 3 | 0 | 3 |
| RRAGB | ZNF649 | 0.750018 | 3 | 0 | 3 |
| RRAGB | KCTD10 | 0.743913 | 3 | 0 | 3 |
| RRAGB | TMEM220 | 0.741426 | 3 | 0 | 3 |
| RRAGB | APOO | 0.741217 | 4 | 0 | 4 |
| RRAGB | BACE2 | 0.739793 | 3 | 0 | 3 |
| RRAGB | LRRC41 | 0.735798 | 3 | 0 | 3 |
| RRAGB | ZNF441 | 0.732379 | 3 | 0 | 3 |
| RRAGB | RERE | 0.731669 | 4 | 0 | 4 |
| RRAGB | UQCRFS1 | 0.728882 | 4 | 0 | 3 |
| RRAGB | LPIN2 | 0.727136 | 3 | 0 | 3 |
| RRAGB | EPM2AIP1 | 0.722578 | 3 | 0 | 3 |
For details and further investigation, click here