| Full name: retinol saturase | Alias Symbol: FLJ20296 | ||
| Type: protein-coding gene | Cytoband: 2p11.2 | ||
| Entrez ID: 54884 | HGNC ID: HGNC:25991 | Ensembl Gene: ENSG00000042445 | OMIM ID: 617597 |
| Drug and gene relationship at DGIdb | |||
Expression of RETSAT:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RETSAT | 54884 | 218124_at | -0.4491 | 0.5278 | |
| GSE20347 | RETSAT | 54884 | 218124_at | -0.7965 | 0.0000 | |
| GSE23400 | RETSAT | 54884 | 218124_at | -0.3933 | 0.0001 | |
| GSE26886 | RETSAT | 54884 | 218124_at | -0.9614 | 0.0000 | |
| GSE29001 | RETSAT | 54884 | 218124_at | -0.7434 | 0.0041 | |
| GSE38129 | RETSAT | 54884 | 218124_at | -0.7176 | 0.0000 | |
| GSE45670 | RETSAT | 54884 | 218124_at | -0.3137 | 0.1081 | |
| GSE63941 | RETSAT | 54884 | 218124_at | -0.3982 | 0.3417 | |
| GSE77861 | RETSAT | 54884 | 218124_at | -0.6469 | 0.0095 | |
| GSE97050 | RETSAT | 54884 | A_23_P209944 | -0.0230 | 0.9262 | |
| SRP007169 | RETSAT | 54884 | RNAseq | -1.3497 | 0.0000 | |
| SRP008496 | RETSAT | 54884 | RNAseq | -1.4463 | 0.0000 | |
| SRP064894 | RETSAT | 54884 | RNAseq | -0.4677 | 0.0142 | |
| SRP133303 | RETSAT | 54884 | RNAseq | -0.5421 | 0.0839 | |
| SRP159526 | RETSAT | 54884 | RNAseq | -0.8312 | 0.0056 | |
| SRP193095 | RETSAT | 54884 | RNAseq | -1.1875 | 0.0000 | |
| SRP219564 | RETSAT | 54884 | RNAseq | -0.5855 | 0.0672 | |
| TCGA | RETSAT | 54884 | RNAseq | -0.3075 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 3.
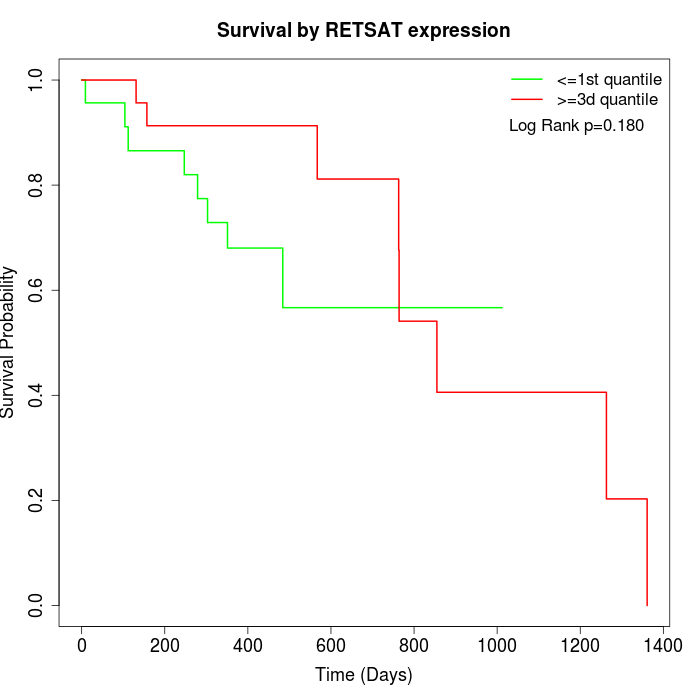
Survival by RETSAT expression:
Note: Click image to view full size file.
Copy number change of RETSAT:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RETSAT | 54884 | 9 | 2 | 19 | |
| GSE20123 | RETSAT | 54884 | 9 | 2 | 19 | |
| GSE43470 | RETSAT | 54884 | 6 | 2 | 35 | |
| GSE46452 | RETSAT | 54884 | 2 | 3 | 54 | |
| GSE47630 | RETSAT | 54884 | 7 | 0 | 33 | |
| GSE54993 | RETSAT | 54884 | 0 | 6 | 64 | |
| GSE54994 | RETSAT | 54884 | 10 | 0 | 43 | |
| GSE60625 | RETSAT | 54884 | 0 | 3 | 8 | |
| GSE74703 | RETSAT | 54884 | 5 | 1 | 30 | |
| GSE74704 | RETSAT | 54884 | 7 | 0 | 13 | |
| TCGA | RETSAT | 54884 | 35 | 3 | 58 |
Total number of gains: 90; Total number of losses: 22; Total Number of normals: 376.
Somatic mutations of RETSAT:
Generating mutation plots.
Highly correlated genes for RETSAT:
Showing top 20/1181 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RETSAT | TRIM7 | 0.783869 | 3 | 0 | 3 |
| RETSAT | C19orf33 | 0.776255 | 3 | 0 | 3 |
| RETSAT | C6orf132 | 0.766041 | 3 | 0 | 3 |
| RETSAT | CERS3 | 0.763472 | 4 | 0 | 4 |
| RETSAT | CAPN14 | 0.761002 | 3 | 0 | 3 |
| RETSAT | PRSS27 | 0.758281 | 3 | 0 | 3 |
| RETSAT | FAR1 | 0.757152 | 3 | 0 | 3 |
| RETSAT | RAB3D | 0.756488 | 4 | 0 | 4 |
| RETSAT | KLHDC8B | 0.75242 | 3 | 0 | 3 |
| RETSAT | CCNYL1 | 0.751832 | 4 | 0 | 4 |
| RETSAT | COMTD1 | 0.749515 | 3 | 0 | 3 |
| RETSAT | SLC16A9 | 0.749005 | 3 | 0 | 3 |
| RETSAT | LYNX1 | 0.741445 | 3 | 0 | 3 |
| RETSAT | TBC1D22B | 0.738736 | 3 | 0 | 3 |
| RETSAT | SMURF1 | 0.737968 | 3 | 0 | 3 |
| RETSAT | BCO2 | 0.737084 | 3 | 0 | 3 |
| RETSAT | CNFN | 0.732026 | 3 | 0 | 3 |
| RETSAT | ACBD3 | 0.730907 | 3 | 0 | 3 |
| RETSAT | DUOXA1 | 0.726239 | 4 | 0 | 4 |
| RETSAT | SLC35A4 | 0.717008 | 3 | 0 | 3 |
For details and further investigation, click here