| Full name: regulator of G protein signaling 13 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 1q31.2 | ||
| Entrez ID: 6003 | HGNC ID: HGNC:9995 | Ensembl Gene: ENSG00000127074 | OMIM ID: 607190 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of RGS13:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RGS13 | 6003 | 210258_at | -0.3927 | 0.8620 | |
| GSE20347 | RGS13 | 6003 | 210258_at | -0.0228 | 0.9120 | |
| GSE23400 | RGS13 | 6003 | 210258_at | -0.0724 | 0.0898 | |
| GSE26886 | RGS13 | 6003 | 210258_at | 0.0269 | 0.8600 | |
| GSE29001 | RGS13 | 6003 | 210258_at | 0.0651 | 0.7070 | |
| GSE38129 | RGS13 | 6003 | 210258_at | -0.4057 | 0.1714 | |
| GSE45670 | RGS13 | 6003 | 210258_at | -1.4512 | 0.0001 | |
| GSE53622 | RGS13 | 6003 | 49863 | -1.2782 | 0.0000 | |
| GSE53624 | RGS13 | 6003 | 49863 | -1.4356 | 0.0000 | |
| GSE63941 | RGS13 | 6003 | 210258_at | -0.2007 | 0.0897 | |
| GSE77861 | RGS13 | 6003 | 1568752_s_at | 0.0144 | 0.8623 | |
| SRP133303 | RGS13 | 6003 | RNAseq | 0.1006 | 0.8230 | |
| TCGA | RGS13 | 6003 | RNAseq | -1.5397 | 0.0018 |
Upregulated datasets: 0; Downregulated datasets: 4.
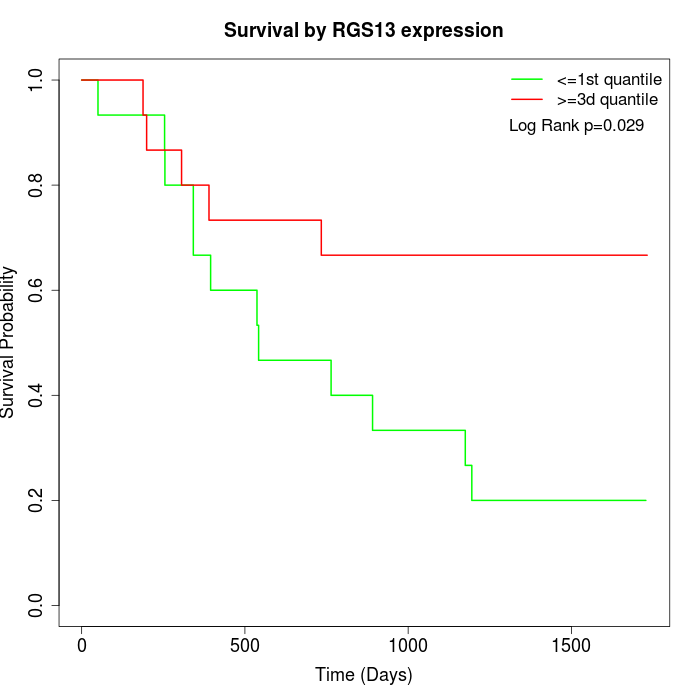
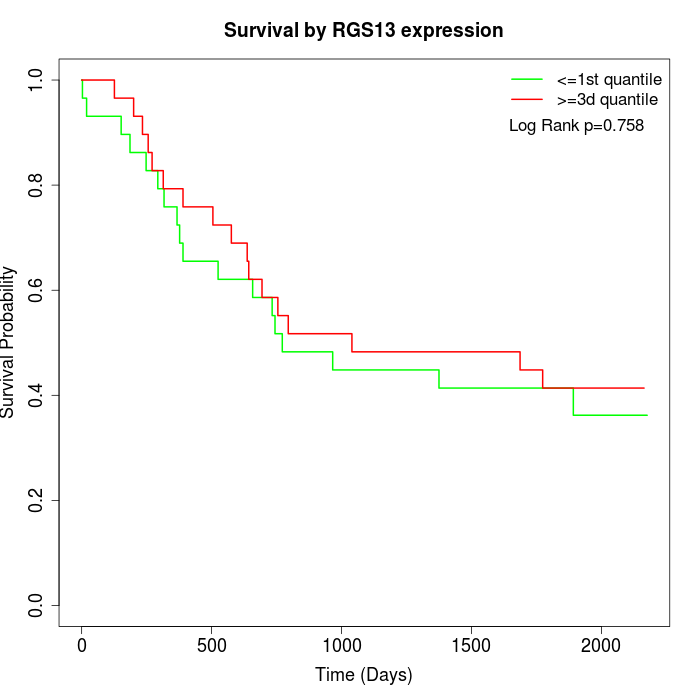
Survival by RGS13 expression:
Note: Click image to view full size file.
Copy number change of RGS13:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RGS13 | 6003 | 9 | 1 | 20 | |
| GSE20123 | RGS13 | 6003 | 9 | 1 | 20 | |
| GSE43470 | RGS13 | 6003 | 5 | 1 | 37 | |
| GSE46452 | RGS13 | 6003 | 3 | 1 | 55 | |
| GSE47630 | RGS13 | 6003 | 14 | 0 | 26 | |
| GSE54993 | RGS13 | 6003 | 0 | 6 | 64 | |
| GSE54994 | RGS13 | 6003 | 15 | 0 | 38 | |
| GSE60625 | RGS13 | 6003 | 0 | 0 | 11 | |
| GSE74703 | RGS13 | 6003 | 5 | 1 | 30 | |
| GSE74704 | RGS13 | 6003 | 3 | 1 | 16 | |
| TCGA | RGS13 | 6003 | 42 | 4 | 50 |
Total number of gains: 105; Total number of losses: 16; Total Number of normals: 367.
Somatic mutations of RGS13:
Generating mutation plots.
Highly correlated genes for RGS13:
Showing top 20/238 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RGS13 | RCSD1 | 0.70855 | 4 | 0 | 4 |
| RGS13 | FLI1 | 0.684207 | 5 | 0 | 5 |
| RGS13 | DAAM2 | 0.681162 | 3 | 0 | 3 |
| RGS13 | TLR10 | 0.671067 | 4 | 0 | 3 |
| RGS13 | PLP1 | 0.636266 | 3 | 0 | 3 |
| RGS13 | CP | 0.628729 | 4 | 0 | 4 |
| RGS13 | LINC00892 | 0.628038 | 3 | 0 | 3 |
| RGS13 | LINC01215 | 0.626483 | 5 | 0 | 5 |
| RGS13 | RGN | 0.625113 | 5 | 0 | 5 |
| RGS13 | TEK | 0.623793 | 5 | 0 | 5 |
| RGS13 | THEMIS | 0.622794 | 3 | 0 | 3 |
| RGS13 | C16orf54 | 0.618955 | 5 | 0 | 5 |
| RGS13 | ICAM2 | 0.616154 | 3 | 0 | 3 |
| RGS13 | RERGL | 0.612162 | 5 | 0 | 5 |
| RGS13 | PTPRN2 | 0.606144 | 4 | 0 | 4 |
| RGS13 | HDC | 0.604379 | 7 | 0 | 5 |
| RGS13 | MMRN1 | 0.601951 | 6 | 0 | 6 |
| RGS13 | GIMAP1 | 0.598226 | 5 | 0 | 4 |
| RGS13 | CNTN3 | 0.596468 | 3 | 0 | 3 |
| RGS13 | PIK3CG | 0.596111 | 5 | 0 | 4 |
For details and further investigation, click here