| Full name: dishevelled associated activator of morphogenesis 2 | Alias Symbol: KIAA0381 | ||
| Type: protein-coding gene | Cytoband: 6p21.2 | ||
| Entrez ID: 23500 | HGNC ID: HGNC:18143 | Ensembl Gene: ENSG00000146122 | OMIM ID: 606627 |
| Drug and gene relationship at DGIdb | |||
DAAM2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04310 | Wnt signaling pathway |
Expression of DAAM2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DAAM2 | 23500 | 212793_at | -1.7817 | 0.1519 | |
| GSE20347 | DAAM2 | 23500 | 212793_at | -0.4765 | 0.0539 | |
| GSE23400 | DAAM2 | 23500 | 212793_at | -0.8477 | 0.0000 | |
| GSE26886 | DAAM2 | 23500 | 212793_at | 0.1925 | 0.4668 | |
| GSE29001 | DAAM2 | 23500 | 212793_at | -0.5138 | 0.0484 | |
| GSE38129 | DAAM2 | 23500 | 212793_at | -1.0994 | 0.0054 | |
| GSE45670 | DAAM2 | 23500 | 212793_at | -2.5385 | 0.0000 | |
| GSE53622 | DAAM2 | 23500 | 12985 | -0.2891 | 0.0274 | |
| GSE53624 | DAAM2 | 23500 | 12985 | -0.1225 | 0.1476 | |
| GSE63941 | DAAM2 | 23500 | 212793_at | -0.3726 | 0.3572 | |
| GSE77861 | DAAM2 | 23500 | 212793_at | 0.1391 | 0.2330 | |
| GSE97050 | DAAM2 | 23500 | A_32_P52785 | -1.4421 | 0.1421 | |
| SRP007169 | DAAM2 | 23500 | RNAseq | 1.4830 | 0.0249 | |
| SRP064894 | DAAM2 | 23500 | RNAseq | -1.0805 | 0.0046 | |
| SRP133303 | DAAM2 | 23500 | RNAseq | -1.0979 | 0.0650 | |
| SRP159526 | DAAM2 | 23500 | RNAseq | -1.6243 | 0.0000 | |
| SRP193095 | DAAM2 | 23500 | RNAseq | 0.3604 | 0.2829 | |
| SRP219564 | DAAM2 | 23500 | RNAseq | -0.5392 | 0.6378 | |
| TCGA | DAAM2 | 23500 | RNAseq | -0.8981 | 0.0000 |
Upregulated datasets: 1; Downregulated datasets: 4.
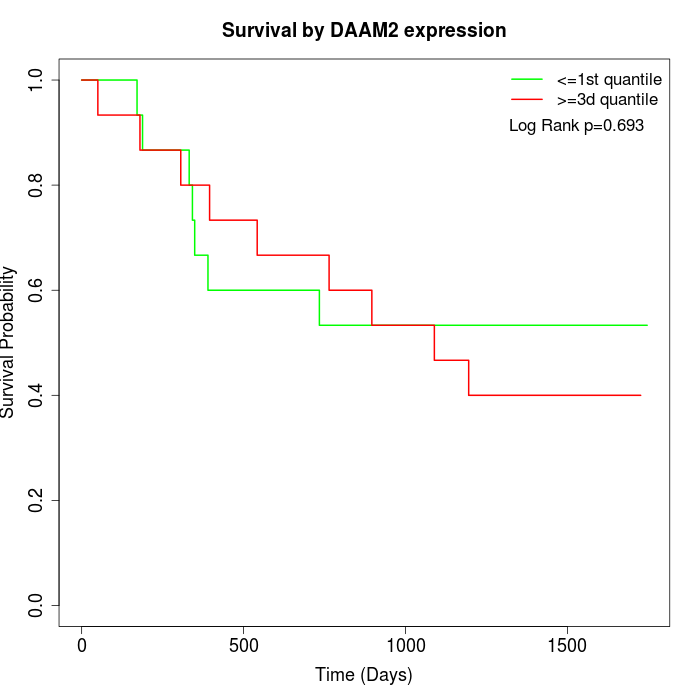
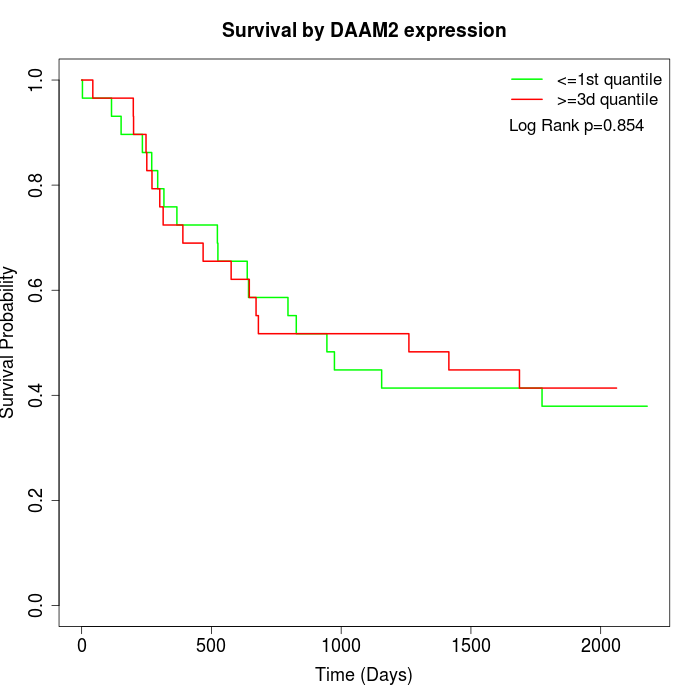
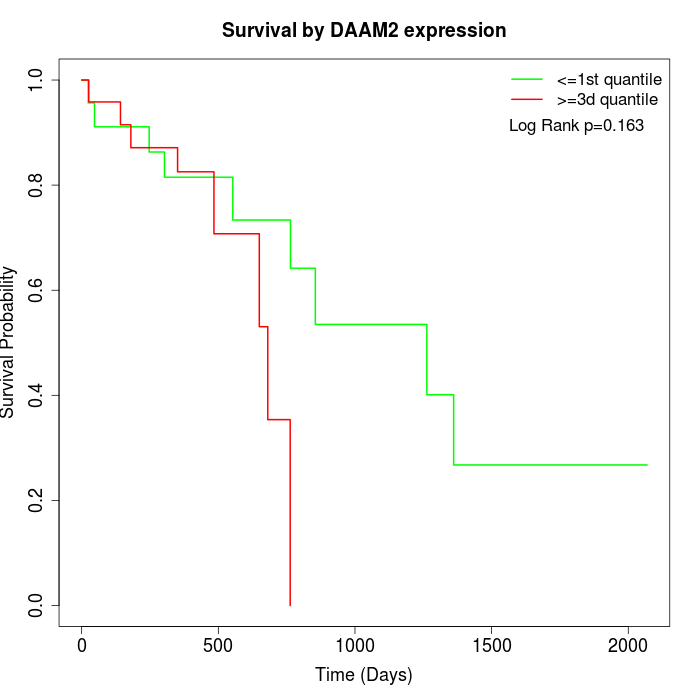
Survival by DAAM2 expression:
Note: Click image to view full size file.
Copy number change of DAAM2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DAAM2 | 23500 | 4 | 1 | 25 | |
| GSE20123 | DAAM2 | 23500 | 4 | 1 | 25 | |
| GSE43470 | DAAM2 | 23500 | 6 | 0 | 37 | |
| GSE46452 | DAAM2 | 23500 | 2 | 9 | 48 | |
| GSE47630 | DAAM2 | 23500 | 8 | 4 | 28 | |
| GSE54993 | DAAM2 | 23500 | 3 | 2 | 65 | |
| GSE54994 | DAAM2 | 23500 | 11 | 4 | 38 | |
| GSE60625 | DAAM2 | 23500 | 0 | 1 | 10 | |
| GSE74703 | DAAM2 | 23500 | 6 | 0 | 30 | |
| GSE74704 | DAAM2 | 23500 | 1 | 1 | 18 | |
| TCGA | DAAM2 | 23500 | 18 | 14 | 64 |
Total number of gains: 63; Total number of losses: 37; Total Number of normals: 388.
Somatic mutations of DAAM2:
Generating mutation plots.
Highly correlated genes for DAAM2:
Showing top 20/1005 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DAAM2 | PPP1R12B | 0.881559 | 8 | 0 | 8 |
| DAAM2 | LMOD1 | 0.851063 | 10 | 0 | 9 |
| DAAM2 | DES | 0.823302 | 9 | 0 | 9 |
| DAAM2 | WWTR1 | 0.801298 | 5 | 0 | 5 |
| DAAM2 | ATP1A2 | 0.801227 | 9 | 0 | 8 |
| DAAM2 | NTN1 | 0.800811 | 3 | 0 | 3 |
| DAAM2 | BOC | 0.79853 | 3 | 0 | 3 |
| DAAM2 | CASQ2 | 0.797949 | 10 | 0 | 9 |
| DAAM2 | OGN | 0.794774 | 8 | 0 | 8 |
| DAAM2 | CHRDL1 | 0.78518 | 10 | 0 | 9 |
| DAAM2 | CILP | 0.776757 | 8 | 0 | 7 |
| DAAM2 | PPP1R12A | 0.774771 | 7 | 0 | 6 |
| DAAM2 | CNN1 | 0.774583 | 11 | 0 | 9 |
| DAAM2 | PLP1 | 0.774378 | 10 | 0 | 9 |
| DAAM2 | TMOD1 | 0.772623 | 9 | 0 | 8 |
| DAAM2 | KCNMB1 | 0.770687 | 11 | 0 | 9 |
| DAAM2 | TGFBR3 | 0.769496 | 6 | 0 | 6 |
| DAAM2 | RASSF3 | 0.764043 | 4 | 0 | 3 |
| DAAM2 | MYH11 | 0.760425 | 9 | 0 | 8 |
| DAAM2 | PRUNE2 | 0.759341 | 11 | 0 | 9 |
For details and further investigation, click here