| Full name: intercellular adhesion molecule 2 | Alias Symbol: CD102 | ||
| Type: protein-coding gene | Cytoband: 17q23.3 | ||
| Entrez ID: 3384 | HGNC ID: HGNC:5345 | Ensembl Gene: ENSG00000108622 | OMIM ID: 146630 |
| Drug and gene relationship at DGIdb | |||
ICAM2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04650 | Natural killer cell mediated cytotoxicity |
Expression of ICAM2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ICAM2 | 3384 | 204683_at | -0.4032 | 0.6423 | |
| GSE20347 | ICAM2 | 3384 | 204683_at | -0.0449 | 0.8206 | |
| GSE23400 | ICAM2 | 3384 | 204683_at | -0.2205 | 0.0033 | |
| GSE26886 | ICAM2 | 3384 | 204683_at | 0.2420 | 0.3684 | |
| GSE29001 | ICAM2 | 3384 | 204683_at | -0.0034 | 0.9842 | |
| GSE38129 | ICAM2 | 3384 | 204683_at | -0.4380 | 0.0916 | |
| GSE45670 | ICAM2 | 3384 | 204683_at | -0.8701 | 0.0019 | |
| GSE63941 | ICAM2 | 3384 | 204683_at | 0.0499 | 0.9065 | |
| GSE77861 | ICAM2 | 3384 | 204683_at | -0.1624 | 0.3341 | |
| GSE97050 | ICAM2 | 3384 | A_23_P152655 | 0.3211 | 0.2317 | |
| SRP007169 | ICAM2 | 3384 | RNAseq | 1.4444 | 0.0260 | |
| SRP064894 | ICAM2 | 3384 | RNAseq | -0.1650 | 0.6649 | |
| SRP133303 | ICAM2 | 3384 | RNAseq | -0.6269 | 0.0126 | |
| SRP159526 | ICAM2 | 3384 | RNAseq | -0.0474 | 0.8917 | |
| SRP193095 | ICAM2 | 3384 | RNAseq | 0.1447 | 0.6818 | |
| SRP219564 | ICAM2 | 3384 | RNAseq | -0.1918 | 0.7316 | |
| TCGA | ICAM2 | 3384 | RNAseq | -0.5011 | 0.0018 |
Upregulated datasets: 1; Downregulated datasets: 0.
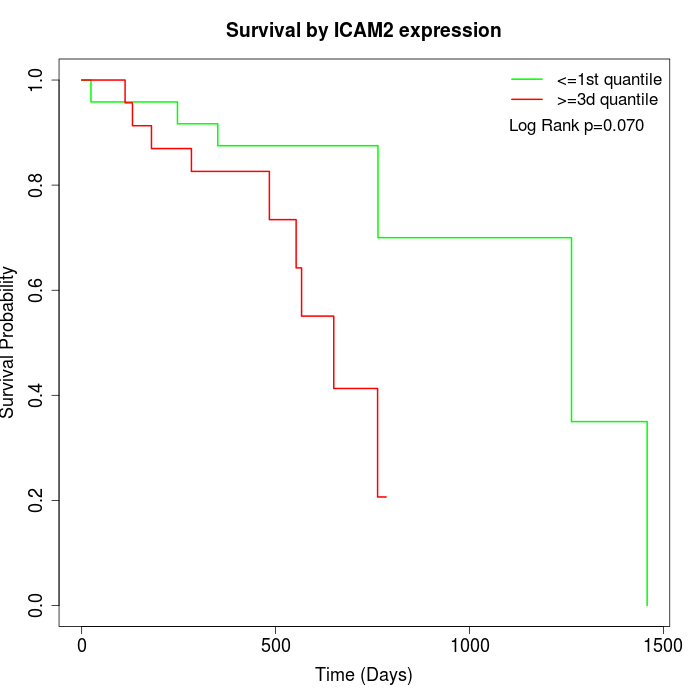
Survival by ICAM2 expression:
Note: Click image to view full size file.
Copy number change of ICAM2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ICAM2 | 3384 | 3 | 1 | 26 | |
| GSE20123 | ICAM2 | 3384 | 3 | 1 | 26 | |
| GSE43470 | ICAM2 | 3384 | 4 | 0 | 39 | |
| GSE46452 | ICAM2 | 3384 | 31 | 0 | 28 | |
| GSE47630 | ICAM2 | 3384 | 7 | 1 | 32 | |
| GSE54993 | ICAM2 | 3384 | 2 | 5 | 63 | |
| GSE54994 | ICAM2 | 3384 | 9 | 4 | 40 | |
| GSE60625 | ICAM2 | 3384 | 4 | 0 | 7 | |
| GSE74703 | ICAM2 | 3384 | 4 | 0 | 32 | |
| GSE74704 | ICAM2 | 3384 | 3 | 1 | 16 | |
| TCGA | ICAM2 | 3384 | 30 | 8 | 58 |
Total number of gains: 100; Total number of losses: 21; Total Number of normals: 367.
Somatic mutations of ICAM2:
Generating mutation plots.
Highly correlated genes for ICAM2:
Showing top 20/478 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ICAM2 | CLEC14A | 0.836515 | 3 | 0 | 3 |
| ICAM2 | RUNX1T1 | 0.756124 | 3 | 0 | 3 |
| ICAM2 | ACKR1 | 0.729638 | 4 | 0 | 4 |
| ICAM2 | ANXA6 | 0.725297 | 6 | 0 | 6 |
| ICAM2 | GNG11 | 0.716391 | 5 | 0 | 4 |
| ICAM2 | FGF7 | 0.713636 | 5 | 0 | 4 |
| ICAM2 | SCG2 | 0.713378 | 4 | 0 | 4 |
| ICAM2 | SLC9A3R2 | 0.709096 | 3 | 0 | 3 |
| ICAM2 | CDH5 | 0.704241 | 6 | 0 | 5 |
| ICAM2 | EBF3 | 0.699636 | 3 | 0 | 3 |
| ICAM2 | ABCA8 | 0.69192 | 4 | 0 | 4 |
| ICAM2 | RASSF2 | 0.691659 | 7 | 0 | 6 |
| ICAM2 | CALHM2 | 0.690076 | 5 | 0 | 5 |
| ICAM2 | CSGALNACT1 | 0.689321 | 3 | 0 | 3 |
| ICAM2 | PIFO | 0.687349 | 3 | 0 | 3 |
| ICAM2 | FILIP1 | 0.686956 | 3 | 0 | 3 |
| ICAM2 | JAM2 | 0.685344 | 5 | 0 | 4 |
| ICAM2 | PRELP | 0.684889 | 5 | 0 | 4 |
| ICAM2 | EID1 | 0.684349 | 3 | 0 | 3 |
| ICAM2 | NLRP7 | 0.684333 | 3 | 0 | 3 |
For details and further investigation, click here