| Full name: phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 7q22.3 | ||
| Entrez ID: 5294 | HGNC ID: HGNC:8978 | Ensembl Gene: ENSG00000105851 | OMIM ID: 601232 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
PIK3CG involved pathways:
Expression of PIK3CG:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PIK3CG | 5294 | 239294_at | 0.3868 | 0.8089 | |
| GSE20347 | PIK3CG | 5294 | 206369_s_at | 0.0069 | 0.9186 | |
| GSE23400 | PIK3CG | 5294 | 206369_s_at | -0.0470 | 0.2309 | |
| GSE26886 | PIK3CG | 5294 | 239294_at | -0.1295 | 0.8084 | |
| GSE29001 | PIK3CG | 5294 | 206369_s_at | -0.0038 | 0.9822 | |
| GSE38129 | PIK3CG | 5294 | 206370_at | -0.0320 | 0.6749 | |
| GSE45670 | PIK3CG | 5294 | 239294_at | -0.5451 | 0.0851 | |
| GSE53622 | PIK3CG | 5294 | 113099 | 0.0120 | 0.9409 | |
| GSE53624 | PIK3CG | 5294 | 113099 | -0.2316 | 0.1609 | |
| GSE63941 | PIK3CG | 5294 | 206369_s_at | 0.1300 | 0.2867 | |
| GSE77861 | PIK3CG | 5294 | 239294_at | -0.0235 | 0.8332 | |
| GSE97050 | PIK3CG | 5294 | A_33_P3304170 | 0.1298 | 0.8735 | |
| SRP007169 | PIK3CG | 5294 | RNAseq | 0.9685 | 0.1425 | |
| SRP008496 | PIK3CG | 5294 | RNAseq | 0.8974 | 0.0720 | |
| SRP064894 | PIK3CG | 5294 | RNAseq | 0.5502 | 0.0517 | |
| SRP133303 | PIK3CG | 5294 | RNAseq | -0.0223 | 0.9014 | |
| SRP159526 | PIK3CG | 5294 | RNAseq | -0.3154 | 0.7193 | |
| SRP193095 | PIK3CG | 5294 | RNAseq | -0.1418 | 0.5730 | |
| SRP219564 | PIK3CG | 5294 | RNAseq | 0.8673 | 0.0735 | |
| TCGA | PIK3CG | 5294 | RNAseq | -0.1504 | 0.5000 |
Upregulated datasets: 0; Downregulated datasets: 0.
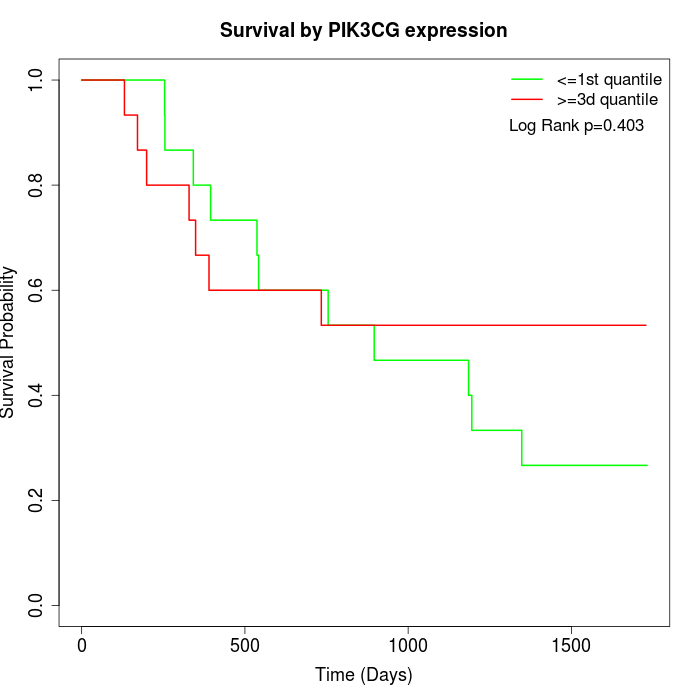
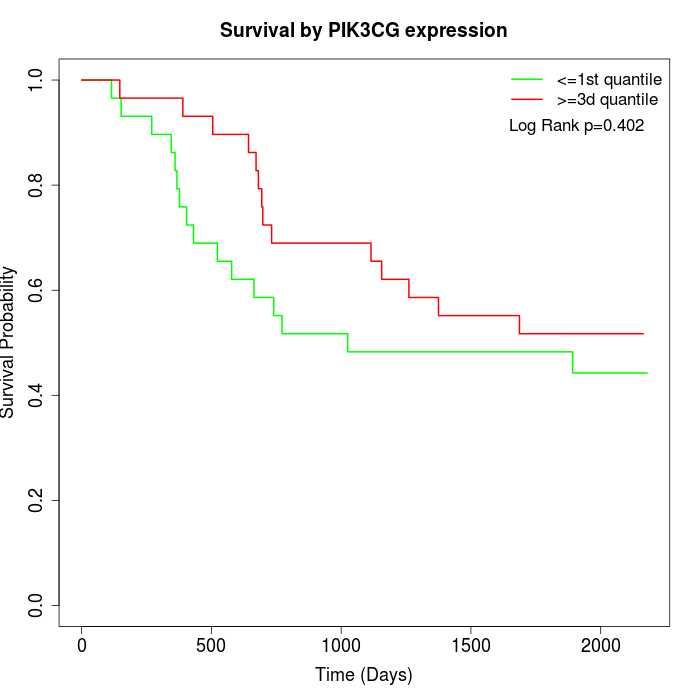
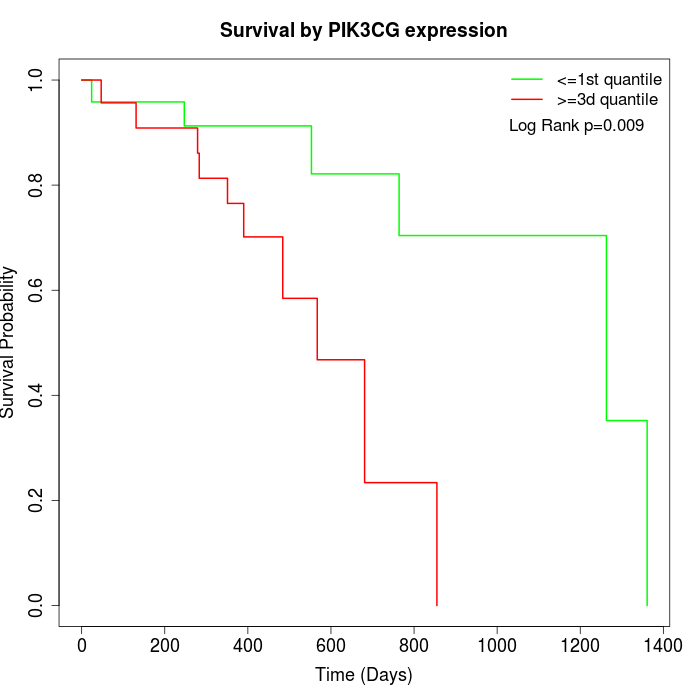
Survival by PIK3CG expression:
Note: Click image to view full size file.
Copy number change of PIK3CG:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PIK3CG | 5294 | 12 | 0 | 18 | |
| GSE20123 | PIK3CG | 5294 | 12 | 0 | 18 | |
| GSE43470 | PIK3CG | 5294 | 6 | 2 | 35 | |
| GSE46452 | PIK3CG | 5294 | 10 | 1 | 48 | |
| GSE47630 | PIK3CG | 5294 | 7 | 3 | 30 | |
| GSE54993 | PIK3CG | 5294 | 2 | 9 | 59 | |
| GSE54994 | PIK3CG | 5294 | 16 | 3 | 34 | |
| GSE60625 | PIK3CG | 5294 | 0 | 0 | 11 | |
| GSE74703 | PIK3CG | 5294 | 6 | 2 | 28 | |
| GSE74704 | PIK3CG | 5294 | 8 | 0 | 12 | |
| TCGA | PIK3CG | 5294 | 51 | 10 | 35 |
Total number of gains: 130; Total number of losses: 30; Total Number of normals: 328.
Somatic mutations of PIK3CG:
Generating mutation plots.
Highly correlated genes for PIK3CG:
Showing top 20/321 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PIK3CG | C16orf54 | 0.834191 | 5 | 0 | 5 |
| PIK3CG | CD1D | 0.786169 | 4 | 0 | 4 |
| PIK3CG | IL10RA | 0.785376 | 7 | 0 | 6 |
| PIK3CG | NCKAP1L | 0.782256 | 7 | 0 | 7 |
| PIK3CG | SLAMF6 | 0.781767 | 4 | 0 | 4 |
| PIK3CG | SNX20 | 0.7805 | 5 | 0 | 4 |
| PIK3CG | GNGT2 | 0.760482 | 3 | 0 | 3 |
| PIK3CG | ARHGAP9 | 0.757638 | 6 | 0 | 5 |
| PIK3CG | IRF8 | 0.750714 | 7 | 0 | 7 |
| PIK3CG | IFNA7 | 0.750691 | 3 | 0 | 3 |
| PIK3CG | CD53 | 0.746342 | 5 | 0 | 5 |
| PIK3CG | ITK | 0.741062 | 8 | 0 | 8 |
| PIK3CG | PARVG | 0.740559 | 5 | 0 | 5 |
| PIK3CG | CD48 | 0.73726 | 8 | 0 | 7 |
| PIK3CG | PPP1R16B | 0.73673 | 4 | 0 | 4 |
| PIK3CG | MPEG1 | 0.7352 | 5 | 0 | 4 |
| PIK3CG | GIMAP2 | 0.731831 | 6 | 0 | 5 |
| PIK3CG | GMFG | 0.730497 | 7 | 0 | 6 |
| PIK3CG | GIMAP6 | 0.723687 | 7 | 0 | 6 |
| PIK3CG | ARHGDIB | 0.722534 | 7 | 0 | 6 |
For details and further investigation, click here